Analysis of Membrane Proteins
Membrane proteins account for about one-third of all proteins expressed in most organisms' genomes and perform some of the most important cellular functions. Due to the complex behavior of membrane proteins in lipid bilayer membranes, the research on biochemistry, biophysics and structure of membrane proteins is still facing great challenges. Nuclear magnetic resonance (NMR) is an advanced tool to study the structure and interaction of membrane proteins at the atomic level.
As an expert in the field of nuclear magnetic resonance, Creative Biostructure can provide customers with membrane protein analysis services by using solution and solid NMR. Our mission is to help customers in membrane protein structure research, dynamics evaluation, folding inspection and membrane protein function research.
Determination of Membrane Protein Structure
Our technologies
| Technology | Technical description | Scope of use | Technical features |
| Solid-state NMR | It is a multifunctional technology, which can be applied to protein samples in almost any state, and is mainly used to analyze insoluble systems, such as membrane proteins and biopolymers. | It is particularly suitable for the study of lipid bilayer intima proteins, especially when the structure, conformation and function are regulated by the composition of the membrane environment. | The application of solid-state NMR is not limited by the size of the studied molecular complex or the viscosity of the medium, because the realization of high resolution does not depend on the reorientation of hot molecules in the solution. |
| Solution NMR | Heteronuclear NMR for soluble proteins | The solution NMR method can be applied to macromolecular complexes experiencing rapid rotational diffusion and correlation time<100ns |
Uniform labeling for solid-state NMR
We usually express proteins in 13C glycerol basic medium for 13C-13C correlation spectroscopy, and in 15N ammonium sulfate for 13C-15N correlation spectroscopy. The 13C-detection experiment is conducted to allocate the spin system, and the heteronuclear experiment is used to sequentially allocate adjacent spin systems.
Uniform labeling for solution NMR
For structural determination, we usually uniformly label proteins with 15N- and 13C- by expressing them in a minimal medium containing 15N ammonium sulfate and 13C glucose as nitrogen and carbon sources. In order to improve spectral resolution, protein side chains are often deuterated by using 2H, 13C glucose instead of 13C glucose. The culture grew in 2H2O, and amide protons were reversely exchanged during protein purification and sample preparation.
Sample preparation
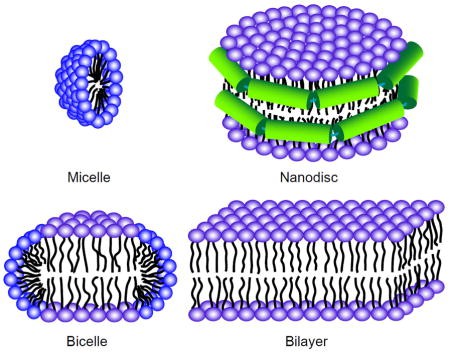 Figure 1. Sample preparation for solution and solid-state NMR of membrane proteins (Liang & Tamm, 2016)
Figure 1. Sample preparation for solution and solid-state NMR of membrane proteins (Liang & Tamm, 2016)
Dynamics Analysis of Membrane Protein
NMR is an excellent method to obtain the specific dynamic information of macromolecular residues, including membrane proteins. NMR provides dynamic information from ps to s to atomic resolution. We can detect the basic biological functions of membrane proteins through extensive NMR technology, such as
- Conformation exchange
- Ligand or inhibitor binding
- Folding and unfolding
- Allosteric regulation of membrane proteins.
Function Analysis of Membrane Protein
Our analysis scope includes but is not limited to
- pH titration
- Small ligand screening
- Protein-protein interactions
- Protein-ligand interactions
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Liang B, Tamm L K. NMR as a tool to investigate the structure, dynamics and function of membrane proteins. Nature Structural & Molecular Biology. 2016. 23(6): 468-474.

