Structure and Dynamics of Membrane-Bound Proteins
In addition to intact membrane proteins, there are also several membrane proteins with functional parts on the membrane surface: peripheral membrane proteins, lipid-anchored proteins and protein parts protruding from the membrane surface. Peripheral membrane proteins temporarily bound to the membrane surface participate in several important cell functions. Because their functional parts exist on the membrane surface in cell signal transduction, cytoskeleton rearrangement and other processes.
The combination of solid and solution NMR experiments is essential to reveal its dynamic structure with significant changes.
As an expert in the field of nuclear magnetic resonance, Creative Biostructure can rely on nuclear magnetic resonance technology to provide customers with structural and kinetic analysis services of membrane binding proteins. Our analysis objects include temporally bound proteins, lipid-anchored proteins and protein motifs processing from the surface and member-bound amyloid proteins.
Our Technologies
The dynamic characteristics of membrane-binding proteins vary greatly between regions where they exhibit structural and dynamic heterogeneity. The correlation time of transmembrane (TM), ring and C-end or N-end protruding from the surface is 10-4-10-2, 10-4-10-5, and<10-6 seconds respectively. Therefore, it is necessary to select the most suitable NMR method to check their respective regions.
To check the rigid area, the technologies we use include dipole-based technology, cross-polarization (CP) magic angle rotation (MAS) and its variants.
To effectively realize heteronuclear correlation and homonuclear correlation, the technologies we use include NCO or NCA correlation based on double cross-polarization (DCP), dipole-assisted rotational resonance (DARR) and its variants. If necessary, we will experiment with low temperatures, which depend on the lipid system used to reduce their movement.
The dipole coupling inevitably attenuates at the moving part protruding from the film surface, and the correlation time is shortened (<10-6s). Therefore, dipole-based technology cannot obtain enough NMR signals. In this case, it is preferable to use monopulse excitation called dipole decoupling (DD) or direct polarization (DP) MAS and scalar-based technology.
To reveal the peptide structure on the membrane surface. Transient NOE (TRNOE) measurement can also be used to reveal the peptide structure on the membrane surface through the NOE data of liquid NMR, provided that the structure is in a fast exchange state between membrane binding and free water solution.
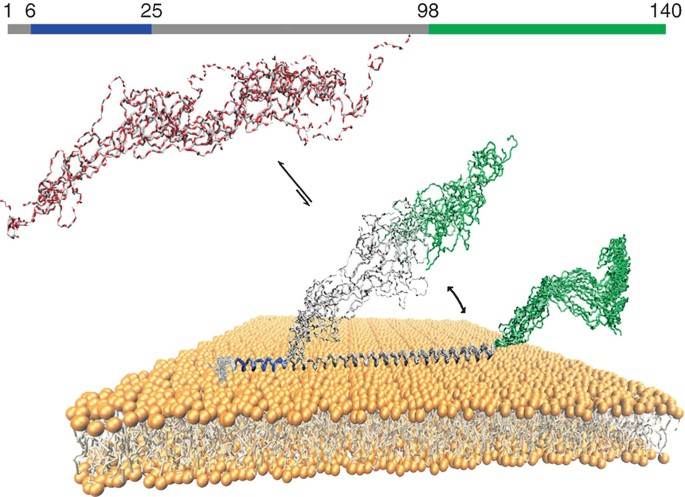 Figure 1. Illustration of the three different regions of αS bound to lipid bilayers (Nishimura et al., 2018)
Figure 1. Illustration of the three different regions of αS bound to lipid bilayers (Nishimura et al., 2018)
Cross-saturation and transfer cross-saturation (TCS) are techniques for identifying the contact residues of peptides and proteins bound to the lipid bait layer.
The chemical exchange saturation transfer (CEST) experiment has been used by us to detect the exchange phenomenon of highly skewed populations in the slow chemical exchange between the visible ground and the invisible excited states of NMR, and to simultaneously monitor the impact on the visible main state peaks in the solution NMR by irradiating the B1 field in each region of the spectrum with weak light.
Subjects of Our Analysis
- Temporally bound proteins - lipid-binding domain
- Lipid-anchored proteins and protein moieties protruding from the surface
- Membrane-bound amyloid proteins
Service Content
We will evaluate the structure and dynamics of membrane-binding proteins through multiple dimensions. Our services include but are not limited to
- Check rigid area.
- Heteronuclear correlation and homonuclear correlation.
- Reveal the peptide structure on the membrane surface.
- Recognition of contact residues of peptides and proteins bound to lipid bait layer.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Nishimura, K., Tanio, M., & Tuzi, S. Structure and dynamics of membrane-bound proteins. Modern Magnetic Resonance. 2018, 669-681.
