Characterization of Protein-RNA Interactions
Understanding the RNA-protein interaction and the structure of the RNA-protein complex is an important research approach. It has been found that the interaction between the ribonucleoprotein (RNP) complex and RNA-protein is the core of many biological processes. Therefore, it is important to analyze the structure of RNA-protein interaction to decipher the function of these RNP complexes and determine the potential target of drug intervention.
Nuclear magnetic resonance spectroscopy is one of the most common technologies in the biophysical tool library, which is used to study the structure, dynamics, and interaction of biomolecules, their ligand complexes, and higher-order molecular assemblies.
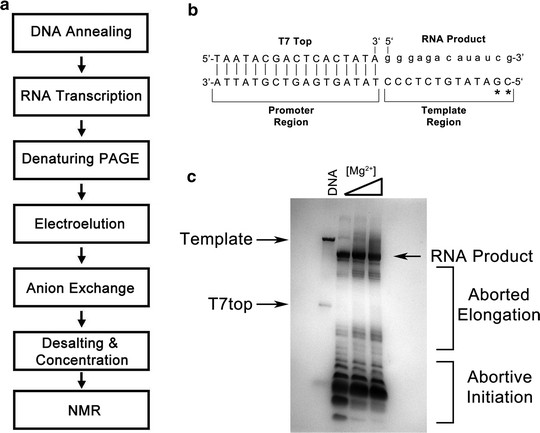 Figure 1. NMR Studies of Protein-RNA Interactions (Theimer et al., 2011)
Figure 1. NMR Studies of Protein-RNA Interactions (Theimer et al., 2011)
As an expert in the field of nuclear magnetic resonance, Creative Biostructure relies on our nuclear magnetic resonance platform to provide our customers with characterization services of protein-RNA interaction. Our services cover topics such as protein and RNA preparation, complex formation, protein-RNA interface recognition, protein and RNA resonance distribution, intermolecular NOE distribution, and structural calculation and analysis. Flexible and comprehensive one-stop service.
Service Process
| Step | Description |
|---|---|
| DNA template design for RNA synthesis | We will design the template according to the actual situation. Of course, you can also provide the template sequence directly. |
| RNA synthesis | All nucleotides are prepared and stored separately so that one or more unlabeled nucleotides can be replaced by 13C- and 15N isotope-labeled nucleotides for the preparation of selectively labeled RNA samples, or all four unlabeled nucleotides can be replaced by 13C- and 15N isotope labeled nucleosides for uniform labeling. |
| RNA purification | Denaturation PAGE is our main method to purify large amounts of RNA into NMR single nucleotide resolution. |
| Preliminary RNA analysis by NMR | After the initial unlabeled RNA sample is prepared, a basic set of NMR experiments will be run, including 1D1H NMR experiments collected in 95% H2O / 5% D2O (5 - 10°C) for pH and salt titration, NOESY collected in 95% H2O / 5% D2O (5 - 10°C) and D2O (20 - 30°C), and TOCSY collected in D2O (20 - 30°C), to evaluate the characteristics of RNA in solution at NMR concentration. The optimum salt and pH conditions are evaluated by comparing the number, intensity, and linewidth of detectable imino proton resonances with the number of imino protons. |
| Analysis RNA–protein Interactions by NMR | Once the chemical shift perturbation method is used to determine several sites on the protein or RNA, other more targeted experiments will narrow the binding region. Finally, the measurement of NOE between RNA and protein provides the most important details for the structural calculation of RNA-protein complex. The filtered and edited NOESY experiment helps to identify NOE used for RNA structure determination and greatly simplifies the recognition of NOE between RNA and protein components of the RNA-protein complex. |
Structural Calculation
To determine the structure of protein and RNA complex by NMR, we will quantify NMR parameters sensitive to molecular conformation, such as
- Dipole relaxation
- Scalar coupling
- Chemical shift
Among them, NOE is the most important because the integration of NOESY cross-peaks is the source of distance constraints used to calculate the structure. The achievable accuracy of distance constraints is limited by the unknown influence of signal-to-noise ratio, peak overlap and molecular mobility, especially at the molecular interface.
We usually use the distance constraints derived from NOE, which are divided into strong, medium and weak categories, and the van der Waals radius is used as the lower limit. The protein dihedral angle limit is usually used in combination with the distance limit in the structural calculation, which can be determined by measuring the scalar coupling or inferred by using the 13C chemical shift. The dihedral angle constraint of the RNA component of the complex can also be measured by scalar coupling.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Theimer, C. A., Smith, N. L., & Khanna, M. NMR studies of Protein-RNA interactions. Methods in Molecular Biology. 2011, 197-218.
