Molecular Dynamics Studies of Pharmaceutically Active Peptides
It is increasingly recognized that peptides and proteins can specifically bind to their targets as well, but because of their small size, they have more drug-like characteristics, so people have a wide interest in peptides as therapeutic agents. Compared with proteins, the smaller size of active peptides translates into lower spectral complexity, making their analysis easier to handle. Modern chemical methods make their assembly and manufacture very easy.
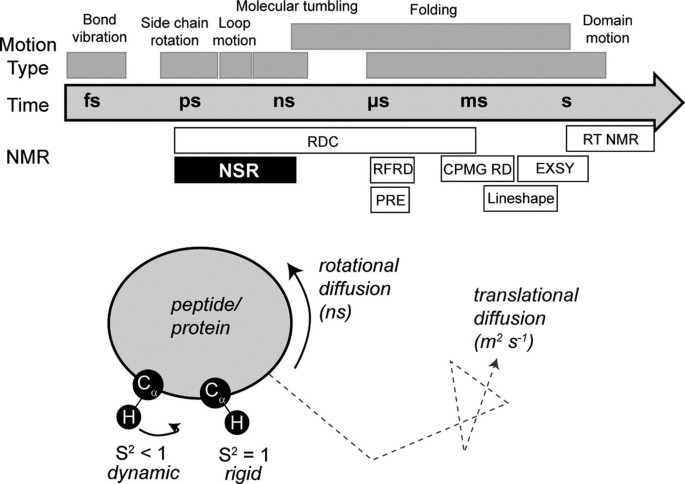 Figure 1. Dynamics and NMR (Wang, 2018)
Figure 1. Dynamics and NMR (Wang, 2018)
As an expert in the field of nuclear magnetic resonance, Creative Biostructure provides NMR relaxation analysis services for drug active peptides to help customers treat active peptides as a better model system for studying and understanding pharmacokinetics.
Determination of Protein Structure by NMR
Nuclear spin relaxation (NSR) is probably one of the most widely used methods with a long history in NMR spectroscopy. It is usually used to determine the range and rate of internal movement on the ps-ns time scale, and monitor basic biophysical phenomena, such as bond vibration and wave, which are considered important for ligand binding and protein conformation.
The transverse relaxation time (T) and self-diffusion coefficient (D) are related to the molecular size. When the small molecule is bound to the large molecule, the T value decreases and the spectral line widens, and the extent of the spectral line widens is linearly related to its binding strength. Compare the spectra of small molecules and proteins before and after binding, and the difference spectrum is the nuclear magnetic signal of small molecules bound to proteins.
Our Workflow of NSR Experiment
In general, NSR programs, the relaxation parameters are first obtained, and then deconvoluted into motion parameters that are easier to interpret. Basically, perform the following steps:
Acquire relaxation observables
- Measure the amplitude or volume of the peak in T1, T2 and hnNOE spectra, preferably under multiple field strengths. Perform three measurements to obtain an error estimate.
- To obtain T1 or T2, the correlation peak intensity of different delay time is fitted to the exponential decay function of the following form: I=I0e-t/T1or2.
- The hnNOE is determined by dividing the peak intensity obtained during proton saturation by the intensity obtained during unsaturation.
Fit relaxation observables to motional parameters
- Qualitative evaluation of data to determine the initial "picture" of the degree of movement.
- Obtain the parameter value of model 1 that is most suitable for the data (i.e., the minimum difference between the experimental and calculated relaxation parameters).
- Repeat the previous step for other models without models.
- Select the parameter values and models that best interpret the data.
After the sample is prepared, the measurement of NSR data is relatively simple. We will collect data under multiple field strengths to increase the number of data points relative to the number of parameters. In addition, we also ensure that the data are obtained in triplicate so that the errors in the data can be estimated. These errors are considered in the fitting process, and then used to estimate the errors in the fitting motion parameters.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Wang, C. K. NMR relaxation analysis of pharmaceutically active peptides. Modern Magnetic Resonance. 2018, 1997-2020.
