NMR Spectroscopy in Fragment-Based Drug Design
Nuclear magnetic resonance (NMR) spectroscopy has developed into a powerful tool for fragment-based drug discovery. Although NMR has traditionally been used to elucidate the three-dimensional structure and dynamics of biological macromolecules and their interactions, it can also be a very valuable tool for reliably identifying small molecules bound to proteins and optimizing lead compounds.
As an expert in the field of fragment-based magnetic resonance, Creative Biostructure provides NMR analysis services for fragment-based drug discovery (FBDD). We provide all NMR analyses involved from fragment library design to structural modeling.
Workflow of Our Services
Our successful fragment-based filtering through NMR depends on the implementation of the following steps
- Get the appropriate fragment library.
- Screening by monitoring changes in protein or ligand spectra.
- Affinity determination.
- Determine the structure of protein fragment complexes and help to produce lead compounds.
Design Fragment Library
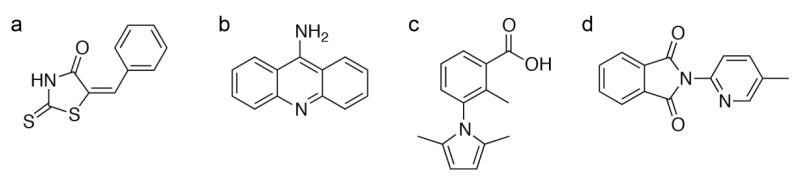 Figure 1. Examples of undesirable molecules in a fragment library. (Harner et al., 2013)
Figure 1. Examples of undesirable molecules in a fragment library. (Harner et al., 2013)
FBDD requires a group of small organic molecules (fragments). The size and quality of the fragment library are crucial to the screening process. For the selection strategy of compounds when constructing a fragment library, we usually follow three rules (molecular weight ≤ 300 Da, ClogP ≤ 3, hydrogen bond donor and acceptor ≤ 3).
Before starting the segment-based screen, we will try to remove the segments that may behave improperly (such as non-specific binding agents, reactive covalent modifiers, chelating agents, or polymerizes).
NMR Fragment-based Screening
Ligand-observed NMR spectroscopy
Due to its reliability, wide applicability and sensitivity, ligand observation NMR spectroscopy has been one of the most commonly used fragment screening methods. The two most commonly used techniques are saturation transfer differential spectroscopy (STD) and Water-LOGSY, both of which are transfer NOE experiments. Both methods are very suitable for detecting weak binding. The larger the target protein, the better its effect.
Other NMR methods for monitoring spectral changes of small molecules include
- Spread edit.
- Experiment based on relaxation.
- Method of using target attached paramagnetic probe (SLAPSTIC).
- Heteronuclear detection scheme (19 F -based screening).
- Target-immobilized NMR screening (TINS).
Screening by chemical shift changes of isotopically labeled proteins
The most robust fragment screening technology is based on monitoring the chemical shift changes of isotope-labeled proteins after adding fragments. One advantage of using a protein signal is that by following the chemical shift perturbation when adding small molecules, not only the hit can be identified, but also the binding strength (Kd value) and binding sites (if the protein signal has been assigned) can be identified.
For proteins larger than 40kDa, we use TROSY-type experiments, such as 1H-15N-TROSY-HSQC, or 1H-13C-HSQC of selectively 13C-labeled (valine, leucine, isoleucine) proteins.
Affinity Determination
We can analyze the NMR titration data to determine the equilibrium dissociation constant (KD) of the fragment binding to its protein target. For this reason, we usually record a series of NMR spectra at fixed protein concentration and increased ligand concentration, and measure the chemical shift perturbation (CSP) of resonance in the spectra. Our methods include protein detection and ligand detection.
Structural Modeling of Protein-ligand Complexes
Our approach includes
- Ligand-observed NMR.
- Protein-observed NMR.
- Chemical shift perturbation mapping.
- NOE-based protein-ligand complexes analysis.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Harner M J, et al. Fragment-based drug discovery using NMR spectroscopy. Journal of Biomolecular NMR. 2013, 56: 65-75.
