Characterization of Protein-Ligand Interactions
The detection and quantitative analysis of protein-ligand interactions are important not only for basic research, but also for the pharmaceutical industry. NMR provides comprehensive information about molecular interactions in solutions with atomic resolution.
As an expert in the field of nuclear magnetic resonance, Creative Biostructure provides customers with characterization services of protein-ligand interactions through ligand-based methods and protein-based methods, depending on the solution NMR.
Justifications for Characterization of Protein-Ligand Interactions
- To obtain structural characteristics of protein-ligand interaction (such as binding site and structural recombination caused by binding).
- To obtain strength information of combination and mechanism of interaction.
- To obtain complete spatial structure and dynamics information of protein-ligand complex (including chemical shift, relaxation rate, spectral peak intensity and coupling constant, etc.).
Ligand-Based Characterization Service for Protein-ligand Interactions
Our experimental technology
- Saturation transfer difference (STD)
- Water-ligand observed via gradient spectroscopy (WaterLOGSY)
- Diffusion-ordered spectroscopy (DOSY)
- Difference of inversion-recovery rate with and without target irradiation (DIRECTION)
- Inter-ligand NOE for pharmacophore mapping (INPHARMA)
- Inter-ligand nuclear Overhauser experiment (ILOE)
- Transferred NOE (trNOE)
All these experiments focused on the changes in NMR signals derived from ligands, thus eliminating the need for the preparation of isotope-labeled proteins.
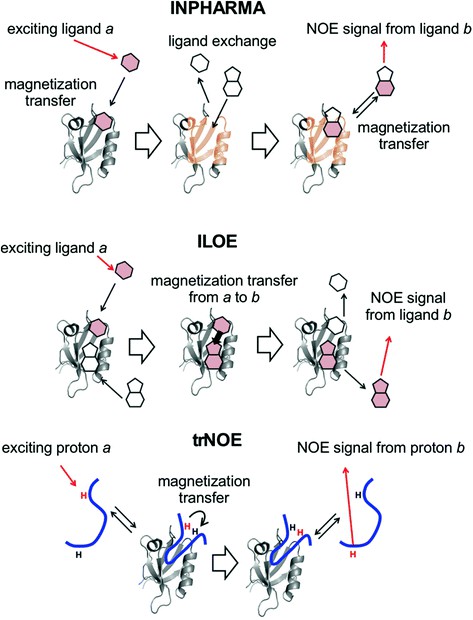 Figure 1. Basic principles of the operations of the ligand-based NMR experiments, INPHARMA, ILOE, and trNOE, for analyzing protein-ligand interactions. (Hiroaki & Kohda, 2017)
Figure 1. Basic principles of the operations of the ligand-based NMR experiments, INPHARMA, ILOE, and trNOE, for analyzing protein-ligand interactions. (Hiroaki & Kohda, 2017)
Problems that we can solve
| Goal | Description | Correspondence method |
| Ligand screening | NMR experiment is combined with a new drug discovery strategy, called fragment-based drug discovery (FBDD). FBDD method found one or several low molecular weight compounds, called fragments, which have weak binding power with the target protein. | STD, WaterLOGSY |
| Pharmacophore determination | Between the lead discovery of drug discovery and the lead optimization process, experiments should be conducted to determine the common core substructure of related compounds with different affinities to the target protein. | DIRECTION, INPHARMA |
| Structural information-driven ligand design | Experiments based on NOE provide useful structural constraints for designing novel molecules with higher affinity for target proteins. | ILOE, trNOE |
Protein-Based Characterization Service for Protein-ligand Interactions
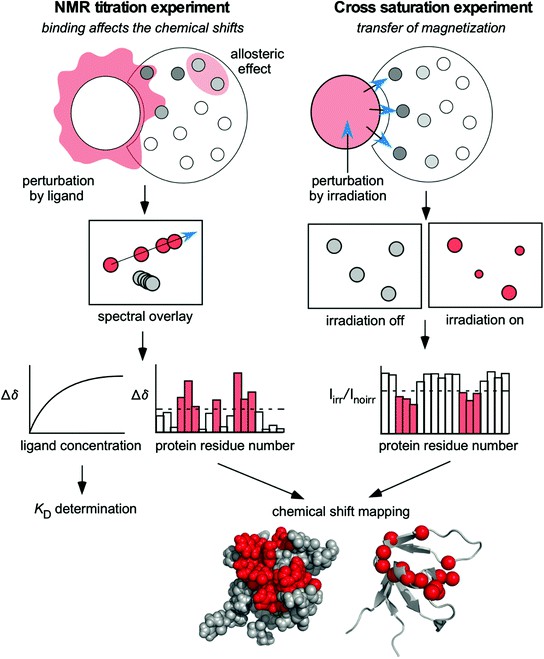 Figure 2. Basic principles of the operations of the protein-based NMR experiments, the NMR titration (alias, chemical shift perturbation) experiment and the cross-saturation experiment. (Hiroaki & Kohda, 2017)
Figure 2. Basic principles of the operations of the protein-based NMR experiments, the NMR titration (alias, chemical shift perturbation) experiment and the cross-saturation experiment. (Hiroaki & Kohda, 2017)
Our experimental technology
Protein-based methods require isotopic labeling of protein samples. After introducing a suitable stable isotope (15N, 13C, or both), measure heteronuclear correlation spectra, such as heteronuclear single quantum coherence (HSQC).
Objectives of the protein-based approaches
- Screening ligands with weak binding to target.
- Obtain information on designing new lead compounds for FBDD.
- Recognize the interface on the target protein molecule.
- Detection of conformational changes of target protein after ligand binding.
- Determine the dissociation constant KD.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Hiroaki, H., & Kohda, D. Protein-ligand interactions studied by NMR. Experimental Approaches of NMR Spectroscopy. 2017. 579-600.
