Characterization of Photoreceptor Membrane Proteins
Rhodopsin is a large class of integrated membrane proteins with photochemical activity. They use retinol as the chromophore. They consist of seven alpha spirals surrounding a transport pathway in which the retina binds via the Schiff base on the conserved lysine located on the seventh helix (helix G). Rhodopsin is divided into two different families.
- Type 1 Rhodopsin is widely distributed in prokaryotes and lower eukaryotes and plays a role as a light-driven ion transporter or light sensor.
- Type 2 Rhodopsin includes photoreceptors in animal eyes.
As an expert in the field of nuclear magnetic resonance, Creative Biostructure provides customers with characterization service of type 1 and type 2 photoreceptor membrane proteins.
Characterization Service of Type 1 Rhodopsin
Our technology - SSNMR
We use solid-state NMR to provide customers with characterization services of microbial rhodopsin. We use the MAS technique to eliminate anisotropic interaction and apply RF pulses to "recouple" anisotropy for structural measurements. The structure, solid-state NMR and function of membrane proteins are sensitive to the lipid membrane simulants used. Compared with other structural methods, solid-state NMR has fewer restrictions from the perspective of sample preparation. It can be applied to membrane proteins recombined in lipids under biochemical conditions very similar to cell membranes.
Our characterization objects
- Bacteriorhodopsin
- Proteorhodopsin
- Sensory Rhodopsins
- Anabaena Sensory Rhodopsin
- Channelrhodopsins
Our characterization services
| Item | Description | Labeling method |
| Chemical shifts | The chemical change is the main reporter of structure and chemical environment. In solid-state NMR, we mainly detect the chemical shifts of carbon or nitrogen atoms, which need to be labeled selectively or uniformly. | In Rhodopsin, our most commonly used marker sites include 15N of the Schiff base (SB) that forms lysine ζ. |
| Distance and torsion angle measurements | The internuclear distance and twist angle represent directly interpretable structural constraints, which can be measured in selectively isotope-labeled proteins to detect changes in protein conformation or structure. | Selective labeling, carbon-carbon and carbon-nitrogen distances can be measured with high accuracy. |
| Complete structure determination | The unified labeling provides the advantage of observing multiple protein sites in a single experiment, thus significantly increasing the information content. | Homogeneous labeling of 13C and 15N can be achieved by heterologous expression of proteins in bacteria or yeast hosts. |
| Solvent accessibility | By observing the enhancement effect of paramagnetic relaxation caused by paramagnetic ions preferentially assigned to the solution, we detect the accessibility to the solvent at a specific location. |
Characterization Service of Type 2 Rhodopsin
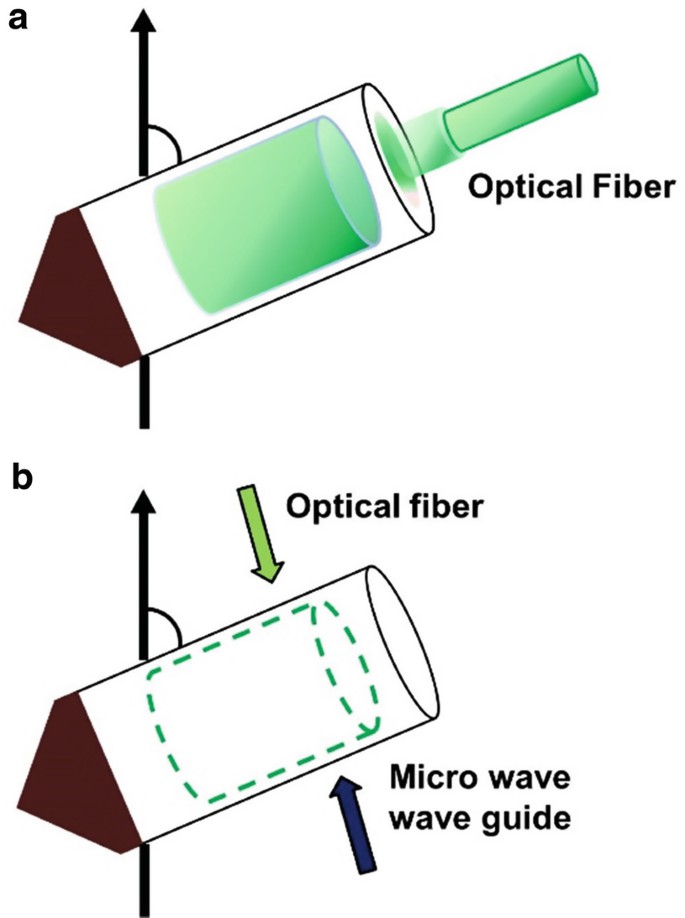 Figure 1. Schematic illustration of in situ photoirradiation apparatus for solid-state NMR spectrometer (Naito et al., 2018)
Figure 1. Schematic illustration of in situ photoirradiation apparatus for solid-state NMR spectrometer (Naito et al., 2018)
Our technology
In situ light irradiation solid-state nuclear magnetic resonance. Photoirradiation solid-state NMR in situ photoirradiation solid-state NMR spectroscopy is particularly suitable for the study of photoreceptor retinal binding membrane proteins, the detection of light intermediates, and the elucidation of light reaction cycle and light-activated structure changes.
Dynamic nuclear polarization (DNP) is a signal enhancement technique that requires paramagnetic labeled samples. Usually, nitrogen and oxygen radicals are introduced into the sample exogenously, and then the SSNMR signal is enhanced by transferring large electron spin polarization to nuclear spin.
Our characterization objects
- Rhodopsin I (SRI)
- Sensory rhodopsin II (SRII)
Our services
- Determining the photoreaction path of SRI using in situ photoirradiation solid-state NMR spectroscopy.
- Photoreaction cycle of pharaonis phoborhodopsin (SRII) determined using in situ photoirradiation solid-state NMR spectroscopy.
- Photoisomerization of Channelrhodopsin-2 determined using photoirradiation DNP SSNMR spectroscopy.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Naito A, Makino Y, Kawamura I. In-situ photo irradiation solid-state NMR spectroscopy applied to retinal-binding membrane proteins. Modern Magnetic Resonance. 2nd edn. Springer, Berlin, 2018: 537-557.
