Cryo-EM for Protein-Ligand Complexes
All living organisms are dependent on two essential biopolymers for their biological functions: nucleic acids and proteins. Nucleic acids (refer to DNA and RNA) are competent to encode and store genetic information in the nucleus of every living organism on Earth. Like the nucleic acids, proteins also are essential parts of organisms and participate in virtually every process within cells. Proteins have the ability to bind other molecules specifically and tightly, such as nucleic acids, other proteins as well as small-molecule substrates and so on. The interaction of biomacromolecules is the fundamental basis of cellular function. Thus it is very important for understanding the biological functions of proteins that studying the interaction of proteins and ligands.
Discovering the tertiary/quaternary structure of proteins and its complexes can provide significant theory basis about how the protein exactly performs its function. X-ray crystallography is the common experimental method of structure determination and it requires proteins that can be easily subjected to crystallize. But For many biological macromolecules, especially biological macromolecular complexes, they are asymmetry, which makes it is difficult to form a spiral symmetry or orderly arranged crystal structure. Cryo-Electron Microscopy (Cryo-EM) is an emerging and expanding structural biology technique. With the new generation of electron detectors and advanced image processing, Cryo-EM has recently undergone a quantum leap progression in resolution and improved its structure determination by single-particle analysis. Without crystallization and only needing a small amount of samples in solution, Cryo-EM can be used to study the dynamic progress of functional complexes, which are often unstable, flexible, transient in their native environments. Due to it can provide a high-resolution tomography and three-dimensional (3D) reconstruction in near-atomic level, Cryo-EM has become a powerful tool for the structural research of biological macromolecular complexes, such as protein-DNA/RNA complexes and protein-ligands, etc.
At Creative Biostructure, we have a wealth of experience in studying the structures of biological macromolecules and its complexes by utilizing Cryo-EM, such as the study of proteins, DNA samples, ribosomes, antigen-antibody complex, and so on. In fact, there has been reported many Cryo-EM structures of biological macromolecules and its complexes. For example, eukaryotic translation complexes provides an abundance of biological insights for understanding the biological progress of translation.
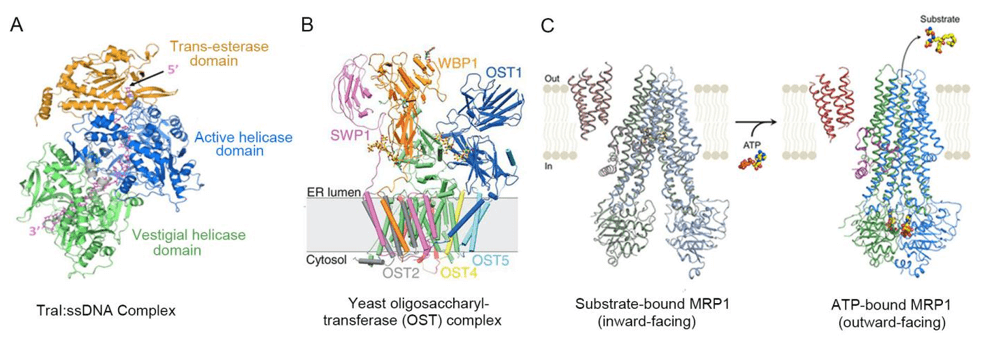 Figure 1. Cryo-EM structures of Protein-Ligand complexes.
Figure 1. Cryo-EM structures of Protein-Ligand complexes.
Creative Biostructure promises to work closely with our customers to provide excellent services of structural studies with unique biological insights.
Please feel free to contact us for a detailed quote.
Ordering Process
References
- Eva Nogales, Sjors H.W. Scheres, Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity. Mol Cell, 2015; 58:677-689.
- Aravindan Ilangovan, Christopher W.M. Kay, et al. Cryo-EM Structure of a Relaxase Reveals the Molecular Basis of DNA Unwinding during Bacterial Conjugation. Cell, 2017;169:708-721.
- Rebekka Wild, Julia Kowal, et al. Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation. Science, 2018; 359:545-550.
- Zachary LeeJohnson, JueChen, ATP Binding Enables Substrate Release from Multidrug Resistance Protein 1. Cell, 2018; 172:81-89.

