NMR Mapping of Protein Interactions in Living Cells
An exceptional asset to cutting-edge biophysical research at Creative Biostructure is offered through our state-of-the-art Nuclear Magnetic Resonance (NMR) platform. Intracellular protein-protein interactions form the basis of most biological processes. Structural aspects of these reactions can now be analyzed in living prokaryotic cells and in atomic detail using NMR spectroscopy technology provided by Creative Biostructure.
To allow the study of biomolecules in their natural environment (that is, in cells), recent attempts have aimed to develop new in vivo techniques for structural biology and cellular imaging. X-ray crystallography and high-resolution Electron Microscopy (EM) are intrinsically excluded from in vivo approaches because of their requirement for pure samples and crystalline or vitrified specimen. In-cell NMR spectroscopy (STINT-NMR), as the only other method for the structural investigation of biomolecules at the atomic level, allows for the direct observation of ‘NMR-active’ atomic nuclei in any NMR-distinguishable environment and can thus be used to investigate labeled proteins structurally in vivo and inside cells.
In-cell NMR has been used to detect protein–protein interactions inside E. coli cells as well as the behavior of intrinsically disordered proteins. In eukaryotic cells, in-cell NMR studies have been performed by injecting proteins into Xenopus laevis oocytes or eggs and, more recently, cell-penetrating peptides have been used to deliver proteins that can be observed in living human cells.
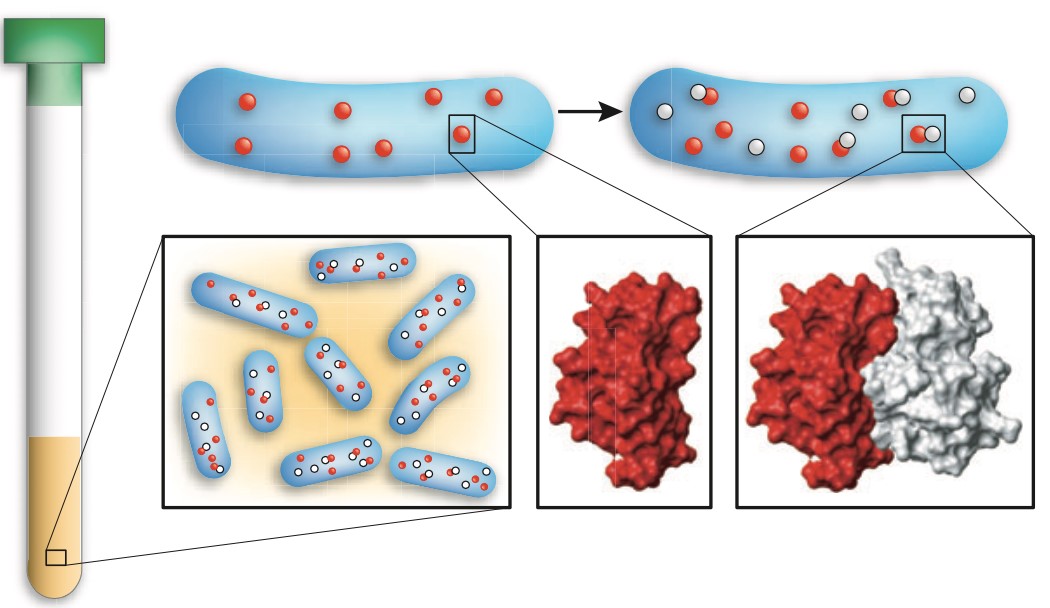 Figure 1. In-cell NMR spectroscopy of protein-protein interactions. To analyze the structural details of a given protein-protein interaction, the two partners are sequentially expressed in E. coli and the suspension of live bacteria is used directly for NMR measurements.
Figure 1. In-cell NMR spectroscopy of protein-protein interactions. To analyze the structural details of a given protein-protein interaction, the two partners are sequentially expressed in E. coli and the suspension of live bacteria is used directly for NMR measurements.
The instrumentation presently available includes four-channel Bruker Avance 500, 600, and 700 MHz NMR spectrometers equipped with cryoprobes. These are state-of-the-art instruments that allow determination of 3D structures of biological macromolecules, including proteins and nucleic acids, both alone and as complexes with various ligands. NMR allows to investigate biomolecular structures and interactions in solution and it also provides information about molecular motions.
Applications of STINT-NMR:
- Structural interpretation of intermolecular, in vivo binding events in a cellular context
- Mutational screens for ligands that are defective in their ability to interact in vivo
- Qualitative in vivo binding affinity analysis
- In vitro measurements when components are difficult to purify or are unstable in their pure form
Creative Biostructure opens new avenues for the investigation of protein conformations at atomic resolution and how they change in response to biological events in living environments. In particular, this approach provides the tools that will permit the effects of molecular crowding in the cytosol, the conformations of proteins that are intrinsically disordered in vitro, and the 3D structures of proteins that are otherwise unstable and difficult to purify to be investigated in living cells.
Given the breadth of applications possible with NMR, and the variety of specific sample requirements, it is advised that you discuss your project with our NMR experts before planning experiments.
Please feel free to contact us for a detailed quote.
Ordering Process
References
- Philipp Selenko & Gerhard Wagner. NMR mapping of protein interactions in living cells. Nature Methods 3, 80 - 81 (2006). doi:10.1038/nmeth0206-80
- Daisuke Sakakibara, et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature. Vol 458, 5 March 2009, doi:10.1038/nature07814

