Characterization of Large Biomolecular Complexes
Large biomolecular complexes are any biological complexes consisting of more than one protein, RNA, DNA, lipid, or carbohydrate molecule. They perform some of the most important processes in the cell.
As an expert in the field of nuclear magnetic resonance, Creative Biostructure uses isotope labeled methyl [13CH3] in the background of complete deuterization, combined with the corresponding methyl TROSY spectrum, to provide customers with the structure, dynamics, and interaction analysis services of large biomolecular complexes.
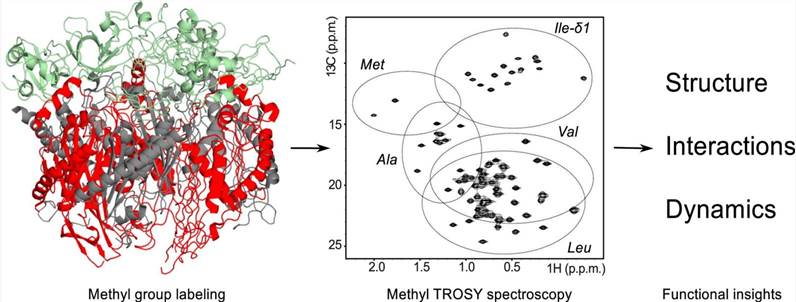 Figure 1. Methyl TROSY spectroscopy: A versatile NMR approach to study challenging biological systems (Schütz S & Sprangers, 2020)
Figure 1. Methyl TROSY spectroscopy: A versatile NMR approach to study challenging biological systems (Schütz S & Sprangers, 2020)
Technical Route and Features
Transverse relaxation optimized spectroscopy (TROSY) Methyl transverse relaxation optimized spectroscopy can determine the structure of proteins up to 80 kDa and analyze proteins up to 900 kDa. The combination of isotope-labeled methyl [13CH3] with the corresponding methyl TROSY spectrum under the background of complete deuterization has been proven to be a successful method for studying high molecular weight complexes.
This strategy benefits from the excellent relaxation properties of methyl, in which its three symmetrically arranged protons and rapid rotation around its triple axis of symmetry produce highly sensitive and well-resolved NMR signals. Methyl is usually located in the hydrophobic interior and the binding surface of proteins, which makes them an important reporter of structural integrity, conformational change, dynamics and interaction.
Methyl Group Labeling Schemes
Our principles of labeling scheme design
The optimal isotope labeling essentially involves finding the correct balance between the number, distribution, and/or location of NMR active nuclei (15N, 13C, 1H) to provide sufficient information about the system, and at the same time, it does not affect the quality and manageability of the recorded spectra with too many NMR active nuclei, otherwise it will lead to signal overlap and broadening.
Our approach
The protein is deuterated uniformly, and the labeled methyl (13CH3) group is selectively introduced into specific amino acids. Hydrogen (1H) is naturally rich in protein, which causes extensive relaxation of transverse magnetization through interaction with each other and with other NMR active nuclei (15N, 13C), thus reducing sensitivity and resolution.
Methyl Group Resonance Assignments
The detailed analysis of NMR spectroscopy requires the allocation of resonance to the corresponding residues in the protein.
| Our methods | Description |
| Divide and conquer | Break the high molecular weight system into smaller building blocks. |
| Mutations to assign methyl groups | Methyl resonance can be distributed by mutagenesis. A single amino acid residue containing a labeled methyl group is replaced by a substituted residue. By comparing the respective spectra before and after mutagenesis, signals belonging to specific methyl groups can be easily identified. |
| Structure-based assignment methods | The methyl distance information obtained from NOE data can be mapped to the known distance from the structure, thus allocating methyl resonance. |
Applications
- Analysis of intermolecular interactions
- Analysis of methionine scanning
- Analysis of protein dynamics
- Analysis of elucidating the complex structure
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
Reference
- Schütz S, Sprangers R. Methyl TROSY spectroscopy: A versatile NMR approach to study challenging biological systems. Progress in Nuclear Magnetic Resonance Spectroscopy. 2020. 116: 56-84.
