Membrane Protein Synthesis and Transport
Membrane proteins play critical roles in various cellular processes, such as signaling, transport, and cell-cell interactions. Their biogenesis and proper localization are essential for maintaining cellular function and integrity. In addition, we also highlight cutting-edge topics in cell-free membrane protein synthesis.
Membrane Protein Biosynthesis
Ribosomal Synthesis
The biosynthesis of membrane proteins begins in the ribosome, a complex molecular machine dedicated to protein translation. Unlike soluble proteins, membrane proteins have hydrophobic regions that necessitate their co-translational insertion into the lipid bilayer. This process is mediated by the signal recognition particle (SRP) pathway:
- Signal Sequence Recognition: Shortly after the ribosome initiates translation, the nascent polypeptide emerges with a signal sequence—a short, hydrophobic stretch of amino acids.
- SRP Binding: The SRP recognizes and binds to the signal sequence arresting further elongation momentarily.
- SRP-Ribosome Complex Targeting: The SRP-ribosome-nascent chain complex is targeted to the SRP receptor on the endoplasmic reticulum (ER) membrane.
- Translocon Engagement: The ribosome docks onto the Sec61 translocon channel, allowing the growing peptide to be inserted into or translocated across the ER membrane.
- Release and Resumption: SRP is released, and translation resumes with the nascent peptide entering the translocon.
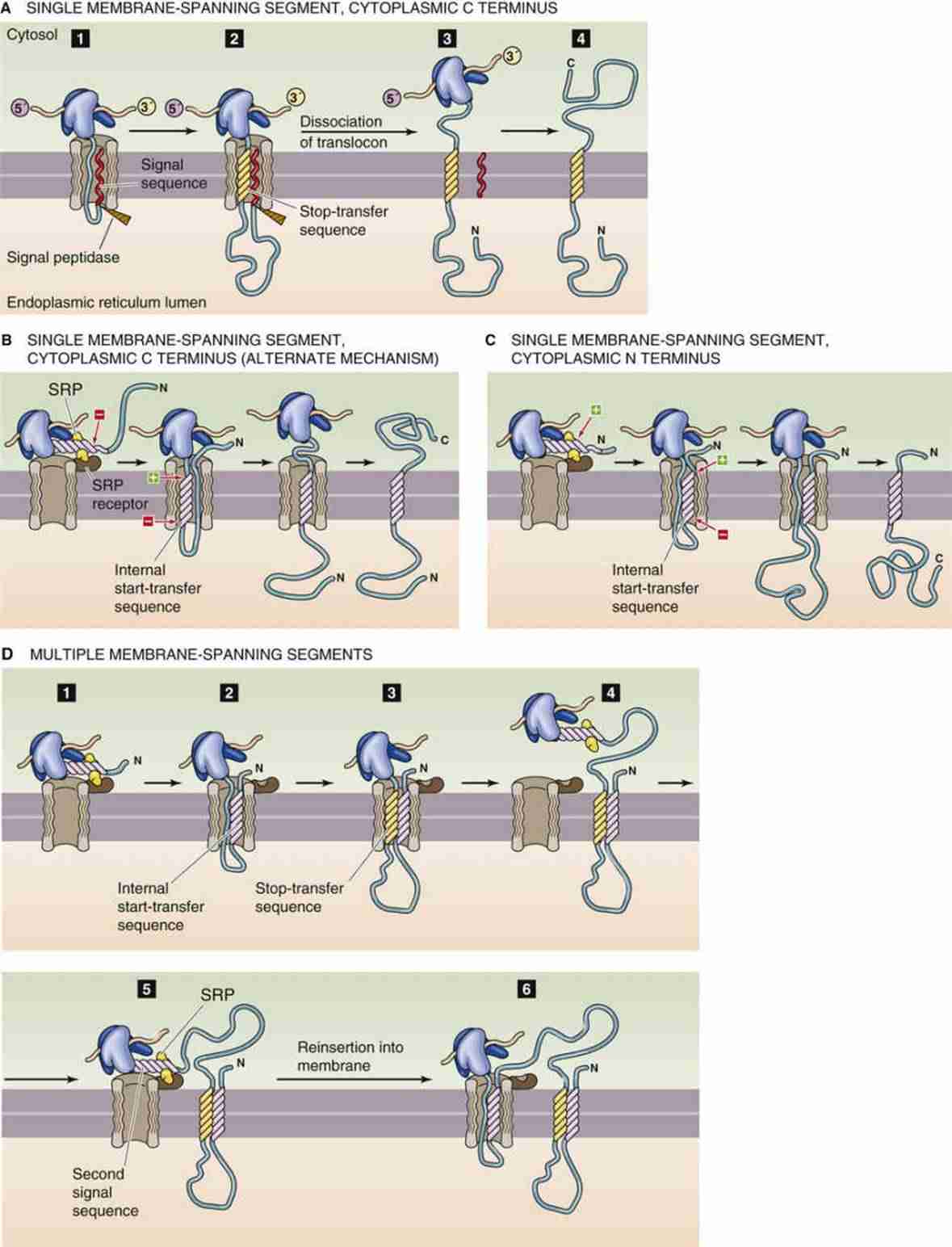 Figure 1. Synthesis of integral membrane proteins. (Michael Caplan)
Figure 1. Synthesis of integral membrane proteins. (Michael Caplan)
ER Membrane Integration
Once the nascent membrane protein enters the ER, it must fold properly and be inserted into the lipid bilayer. The translocon accommodates both lateral exit of transmembrane segments into the lipid bilayer and translocation of luminal domains into the ER lumen. Molecular chaperones and enzymes within the ER, such as BiP and protein disulfide isomerase (PDI), assist in the correct folding of membrane proteins.
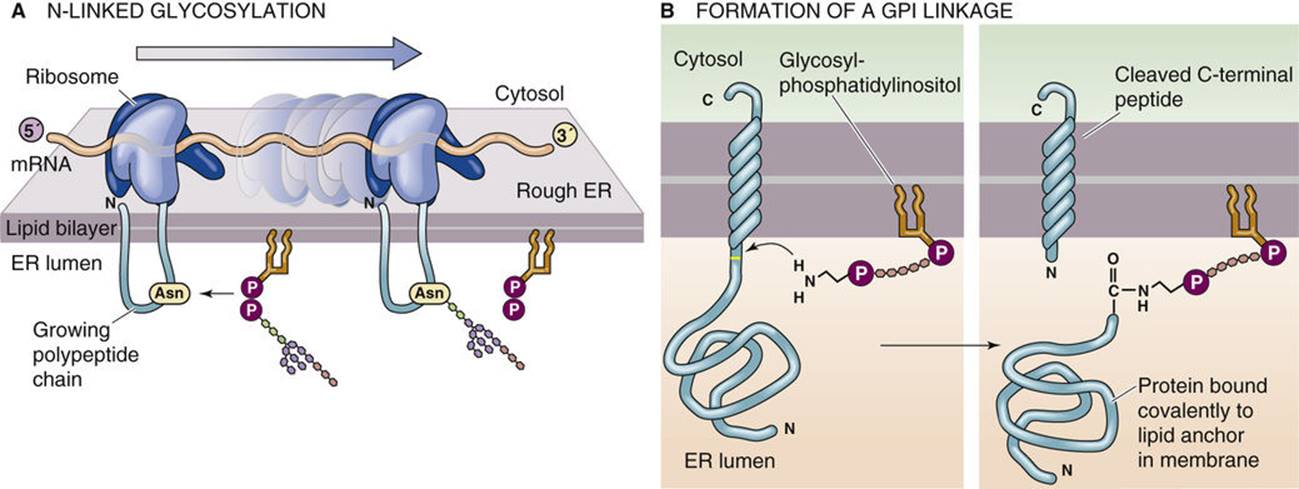 Figure 2. Post-translational modifications of integral membrane proteins. (Michael Caplan)
Figure 2. Post-translational modifications of integral membrane proteins. (Michael Caplan)
Trafficking of Membrane Proteins
Vesicular Transport
After synthesis and initial folding within the ER, membrane proteins are transported to their functional destinations via the vesicular transport system. This system involves several key steps:
- Cargo Recognition: Membrane proteins are selected and packaged into vesicles. Cargo receptors and coat protein complexes (e.g., COPII for ER-to-Golgi transport) play vital roles in this selection.
- Vesicle Formation and Budding: Vesicles bud off from the ER and travel to the Golgi apparatus. Small GTPases, such as Sar1 and ARF, regulate vesicle budding and coat assembly.
- Tethering and Fusion: Vesicles are strategically tethered to their target membranes by tethering factors and subsequently fuse with them through SNARE protein complexes, facilitating cargo delivery.
Golgi Processing and Sorting
Within the Golgi apparatus, membrane proteins undergo further modifications, such as glycosylation, phosphorylation, and sulfation. The Golgi also functions as a critical sorting station, determining the final destination of membrane proteins:
- Route to Plasma Membrane: Proteins destined for the plasma membrane are packaged into vesicles for exocytosis.
- Route to Lysosomes/Endosomes: Proteins targeting lysosomes or endosomes contain specific sorting signals that guide them through the vesicular transport pathways.
Cell-Free Membrane Protein Synthesis
Recent advancements in biotechnology have led to the development of cell-free membrane protein synthesis systems. These systems provide a versatile and efficient platform for synthesizing membrane proteins without the constraints of living cells.
Cell-free membrane protein synthesis leverages cell extracts rich in ribosomes, tRNAs, amino acids, and transcription/translation factors to facilitate protein synthesis outside the cell. The incorporation of synthetic lipid bilayers or nanodiscs into the reaction mixtures ensures proper folding and insertion of membrane proteins.
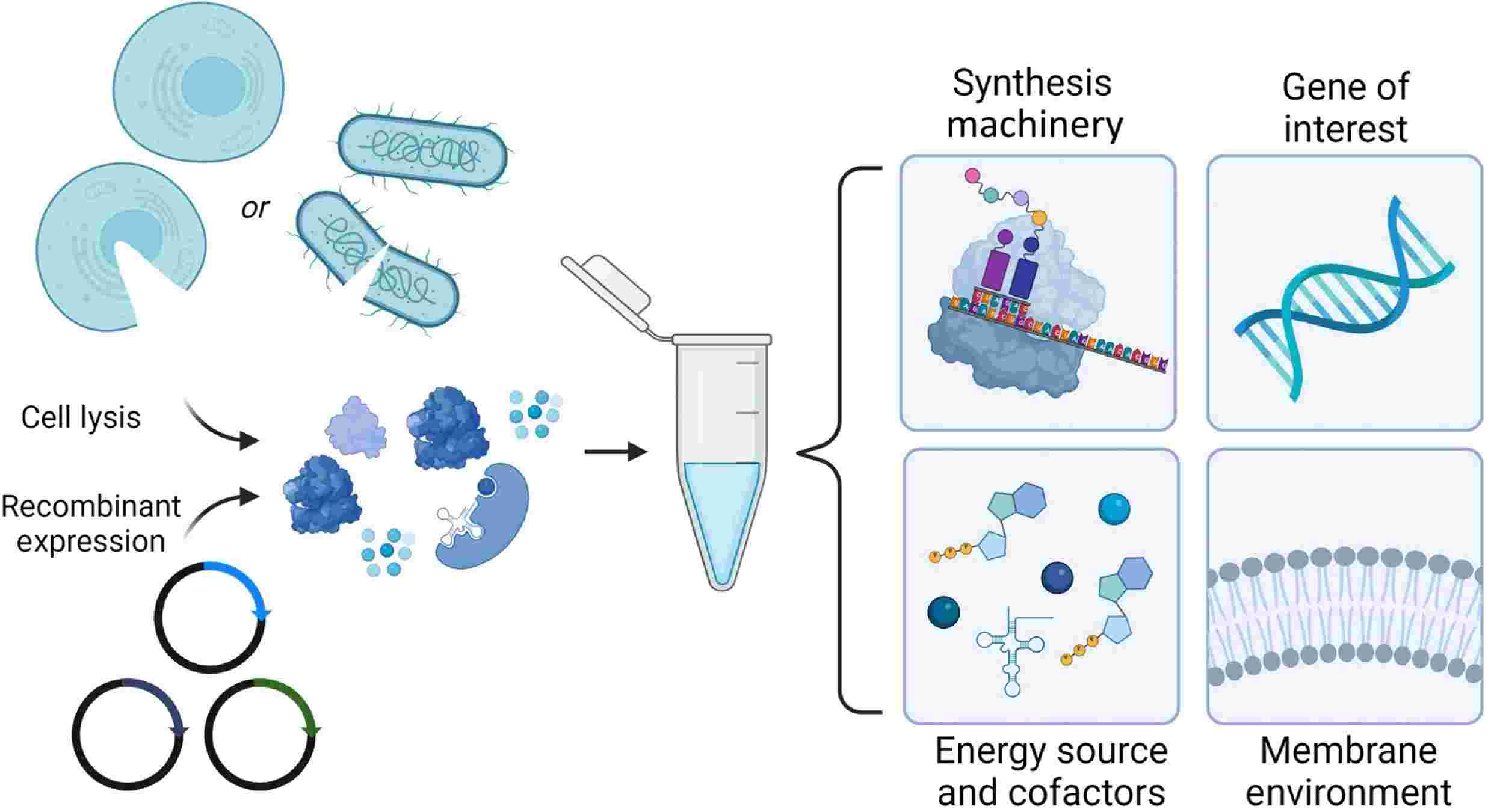 Figure 3. Cell-free membrane protein synthesis. (Manzer Z A, et al., 2023)
Figure 3. Cell-free membrane protein synthesis. (Manzer Z A, et al., 2023)
Advantages of Cell-Free Membrane Protein Synthesis
- Speed and Flexibility: Cell-free membrane protein synthesis allows for rapid synthesis of membrane proteins without the need for cell culture, making it an attractive option for high-throughput studies.
- Simplified Protein Engineering: The open-system nature of cell-free membrane protein synthesis simplifies the manipulation of reaction conditions and incorporation of non-natural amino acids or labels.
- Study of Toxic Proteins: Membrane proteins that are toxic or challenging to express in living cells can be successfully synthesized and characterized using cell-free membrane protein synthesis.
Technical Approaches in Cell-Free Membrane Protein Synthesis
- PURE System: The Protein synthesis Using Recombinant Elements (PURE) system is a reconstituted translation system composed of essential purified components, offering a high degree of control and reduced complexity.
- Wheat Germ Extract: Extracts from wheat germ are commonly used for cell-free membrane protein synthesis due to their high translational capacity and suitability for large-scale protein production.
- Lipid Nanodiscs and SMALPs (Styrene-Maleic Acid Lipid Particles): These lipid-based systems provide a membrane mimetic environment that stabilizes synthesized membrane proteins, facilitating functional and structural studies.
Applications of Cell-Free Membrane Protein Synthesis
- Structural Biology: Cell-free membrane protein synthesis enables the production of membrane proteins for structural analysis via techniques such as X-ray crystallography and cryo-electron microscopy (cryo-EM).
- Drug Screening: The rapid synthesis and functional testing of membrane proteins make cell-free membrane protein synthesis a valuable tool for high-throughput screening of drug candidates.
- Functional Studies: Cell-free membrane protein synthesis allows for the detailed investigation of membrane protein functions, interactions, and dynamics without the background noise from cellular components.
Adcanced Techniques in Membrane Protein Synthesis and Transport Research
| Techniques | Description |
| Nanodisc Technology | Nanodisc technology replicates the cellular membrane environment, enabling membrane proteins to retain their natural shape and function. This is crucial for examining how these proteins interact with drugs, toxins, and other substances. |
| Cell-free Protein Synthesis System | The cell-free protein synthesis system is a powerful tool for expressing, folding, and assembling membrane proteins outside of cells. It simplifies the process by bypassing the complexity of the cell's interior, allowing for direct observation and control of protein synthesis and assembly. |
| Single-Molecule Tracking | This advanced imaging technique enables scientists to see individual membrane proteins moving within living cells. By attaching fluorescent tags to proteins, researchers can track their movement and interactions, offering deep insights into protein trafficking, targeting, and the impact of membrane microdomains on protein function. |
| High-Throughput Screening and Genomics | High-throughput screening and genomics have advanced the discovery of genes and pathways in membrane protein synthesis and transport. Tools like CRISPR-Cas9 and RNAi precisely edit genes to study their roles in trafficking. Combined with next-generation sequencing, these reveal new regulatory networks and trafficking components. |
| Dynamic Endomembrane Organellar Profiling Technology | This technology efficiently separates and identifies proteins from various organelles simultaneously, focusing on those involved in vesicle transport. It surpasses traditional limitations, offering a fresh approach to studying the cell's internal membrane transport system. |
| Computational Modeling | Computational modeling and bioinformatics are essential for studying membrane protein synthesis and transport. Molecular dynamics simulations forecast how these proteins behave and interact in lipid bilayers. Bioinformatics tools analyze extensive proteomic and genomic data, helping to find new trafficking signals and map protein networks. |
Creative Biostructure provides custom membrane protein and cell-free membrane protein synthesis to support your research needs in membrane biology. If you are interested in our solutions, please feel free to contact us for more information.
Reference
- Manzer Z A, Selivanovitch E, Ostwalt A R, et al Membrane protein synthesis: no cells require. Trends in Biochemical Sciences. 2023. 48(7): 642-654.