Structural Research of Receptor Tyrosine Kinase (RTK) Family
Receptor tyrosine kinases (RTK) are single transmembrane receptors that catalyze the transfer of γ-phosphate from ATP to tyrosine residues in protein substrates. Research has shown that RTK dimerization and phosphorylation of specific tyrosine residues in the cytoplasmic fraction are promoted by ligand binding. This enhances RTK catalytic activity and provides docking sites for downstream signaling proteins. RTK plays a critical role in embryogenesis, normal physiology, and a wide range of diseases and has become an essential target for cancer drugs. Therefore, cutting-edge technologies such as X-ray crystallography and cryo-electron microscopy are needed to deepen the understanding of the molecular mechanism of RTK.
Advances in RTK research technology
Further progress in understanding RTK activation will require an understanding of the overall ligand-specific dimerization microstates of each RTK. And while crystal structure-based, biophysical methods such as small-angle X-ray scattering (SAXS) and hydrogen provide critical insights into protein static picture-deuterium exchange mass spectrometry (HXD) to reveal changes in RTK conformation throughout the activation process, it is increasingly possible to apply nuclear magnetic resonance (NMR) to research RTK structures in solution.
Structural analysis of RTK
Using negative staining electron microscopy (NS-EM) and cryo-electron microscopy (cryo-EM), it is possible to observe the conformation of full-length RTK proteins deposited on EM grids. In contrast to non-receptor TK, RTK has an extracellular region at the N-terminal end of the protein containing an extracellular domain for recognition of extracellular signaling molecules, an α-helix that spans the plasma membrane, and a structural domain of the C-terminal tyrosine kinase. RTK undergoes conformational changes, mainly involving dimerization, which leads to activation of the kinase and transmission of the signal into the cell.
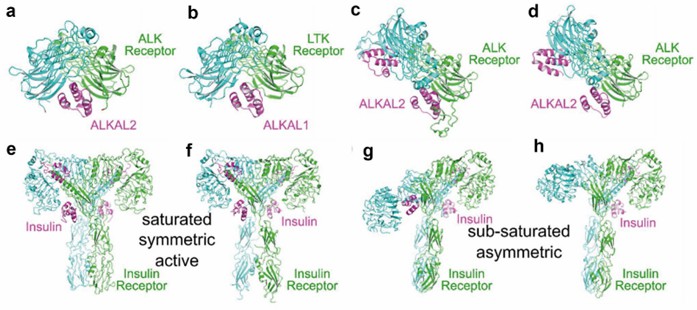 Figure 1. Ligand-bound dimeric RTK structures. (Rygiel KA, Elkins JM, 2023)
Figure 1. Ligand-bound dimeric RTK structures. (Rygiel KA, Elkins JM, 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| The kinase domain of RTKC8 | Monosiga brevicollis | X-ray diffraction | 1.95 Å | 8E4T |
| FGFR1 in complex with ponatinib. | Homo sapiens | X-ray diffraction | 2.12 Å | 4V04 |
| Ephrin B4 (EphB4) receptor protein kinase with Dasatinib | Homo sapiens | Cryo-EM single particle analysis | 1.157 Å | 6FNM |
| Insulin receptor-related receptor's transmembrane domain | Homo sapiens | SOLUTION NMR | / | 7PL4 |
| EphB4 kinase domain inhibitor complex | Homo sapiens | X-ray diffraction | 2.11 Å | 2YN8 |
| Ephrin B4 (EphB4) Receptor Protein Kinase | Homo sapiens | X-ray diffraction | 1.269 Å | 6FNL |
| EphB4 kinase domain with an oxindole inhibitor | Homo sapiens | X-ray diffraction | 2.33 Å | 4AW5 |
| Insulin receptor-related receptor (IRR) in apo-state captured at pH 7 | Homo sapiens | Cryo-EM single particle analysis | 3.5 Å | 7TYK |
| Insulin receptor-related receptor (IRR) in apo-state captured at pH 7 | Homo sapiens | Cryo-EM single particle analysis | 3.3 Å | 7TYJ |
| Insulin receptor-related receptor (IRR) in active-state captured at pH 9 | Homo sapiens | Cryo-EM single particle analysis | 3.4 Å | 7TYM |
| EphB4 in complex with staurosporine | Homo sapiens | X-ray diffraction | 2.5 Å | 3ZEW |
| FGFR1 in complex with AZD4547. | Homo sapiens | X-ray diffraction | 2.57 Å | 4V05 |
| LTK: ALKAL1 complex stabilized by a Nanobody | Homo sapiens | X-ray diffraction | 1.59 Å | 7NX0 |
| dimeric transmembrane domain of the receptor tyrosine kinase EphA1 in lipid bicelles at pH 4.3 | Homo sapiens | SOLUTION NMR | / | 2K1K |
| dimeric transmembrane domain of the receptor tyrosine kinase EphA1 in lipid bicelles at pH 6.3 | Homo sapiens | SOLUTION NMR | / | 2K1L |
| Sam domain from the receptor tyrosine kinase EphB2 | Gallus gallus | SOLUTION NMR | / | 1SGG |
| Extracellular WNT-binding module | Drosophila melanogaster | X-ray diffraction | 1.75 Å | 7ME4 |
| Ephrin B4 (EphB4) Receptor Protein Kinase with NVP-BHG712 | Homo sapiens | X-ray diffraction | 1.468 Å | 6FNI |
| TG domain of LTK | Homo sapiens | X-ray diffraction | 1.3 Å | 7NX1 |
| SAM Domain of Human Ephrin Type-B Receptor 4 | Homo sapiens | X-ray diffraction | 2.1 Å | 2QKQ |
| SAM Domain of Human Ephrin Type-A Receptor 1 (EphA1) | Homo sapiens | X-ray diffraction | 2 Å | 3HIL |
Table 1. Structural research of receptor tyrosine kinase (RTK) family.
Creative Biostructure is a leading provider of biomolecular structural research, offering personalized solutions and flexible collaboration opportunities to meet clients' specific requirements. Whether it is basic structural research or functional analysis of the receptor tyrosine kinase (RTK) family, we provide accurate and reliable results.
With cutting-edge technologies including cryo-electron microscopy (cryo-EM), NMR spectroscopy, and X-ray crystallography, we can provide high-resolution structural analysis services. Our team consists of experienced scientists and researchers solving intricate biological problems. If you require structural research in biomolecules, contact us and our service will help you closer to achieving your scientific goals.
References
- Dyla M, et al. Structure and Mechanism of P-Type ATPase Ion Pumps. Annu Rev Biochem. 2020。89:583-603.
- Bublitz M, et al. Structural studies of P-type ATPase-ligand complexes using an X-ray free-electron laser. IUCrJ. 2015.2(Pt 4):409-420.
- Dyla M, et al. Structural dynamics of P-type ATPase ion pumps. Biochem Soc Trans. 2019。47(5):1247-1257.