Structural Research of Novel Receptors
Membrane receptors are integrated membrane proteins that affect communication between cells and external space through cascade chemical changes in the cell membrane. Membrane receptors are classified into three main groups including ion channel-linked receptors, enzyme-linked receptors, and G protein-coupled receptors. Simple receptor polypeptide chains cross the lipid bilayer once, whereas complex receptors, such as G protein-coupled receptors, cross the membrane up to seven times.
Ion channel-linked receptors
During signal transduction in neurons, neurotransmitters bind to receptors that open ion channels and allow ions to enter the cell, thereby exciting the cell. Such as the acetylcholine receptor, consists of four subunits, α, β, γ, and δ. Its natural state is closed and empty. When an acetylcholine molecule binds to the binding site on the α subunit, the receptor conformation changes, allowing ions and small molecules to enter the cell.
Enzyme-linked receptors
Enzyme-linked receptors have intracellular structural domains associated with enzymes and cell surface receptors. Its binding to extracellular ligands elicits enzymatic activity on the intracellular side. The transmembrane region consists of a single alpha helix region of the peptide chain.
G protein-coupled receptors
G protein-coupled receptors (GPCRs) are found in eukaryotes and cross the cell membrane seven times in six loops that bind to ligands and activate G proteins. The α-subunit of G proteins, together with GTP, can be dissociated from the β- and γ-subunits, which can further affect intracellular signaling proteins or directly target functional proteins. GPCRs have been implicated in a wide range of diseases, and are therefore the targets of many modern drugs.
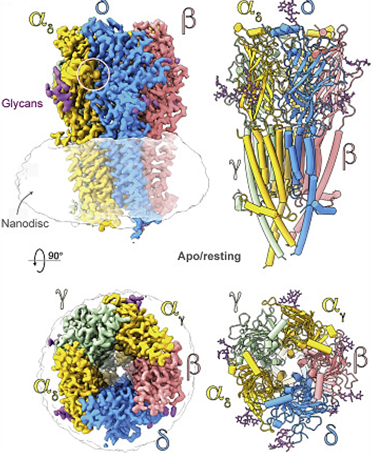 Figure 1. Top and side views of the nicotinic acetylcholine receptor (nAChR) cryo-EM reconstruction. (Zarkadas E, et al., 2022)
Figure 1. Top and side views of the nicotinic acetylcholine receptor (nAChR) cryo-EM reconstruction. (Zarkadas E, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| STX-478, a Mutant-Selective, Allosteric Inhibitor bound to H1047R PI3Kalpha | Homo sapiens | X-ray diffraction | 2.928 Å | 8TGD |
| TMEM16F, with Calcium and PIP2, no inhibitor, Cl1 | Mus musculus | Cryo-EM single particle analysis | 3.2 Å | 8TAL |
| Abl kinase in complex with SKI and asciminib | Homo sapiens | X-ray diffraction | 2.86 Å | 8SSN |
| LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunits TARP gamma-5 and cornichon-2 | Homo sapiens | Cryo-EM single particle analysis | 3.66 Å | 8SSB |
| Therapeutic antibody (favezelimab) bound to human LAG3 local refined | Homo sapiens | Cryo-EM single particle analysis | 3.533 Å | 8SR0 |
| Therapeutic antibody (favezelimab) bound to human LAG3 | Homo sapiens | Cryo-EM single particle analysis | 3.61 Å | 8SO3 |
| TIR-1 protein | Caenorhabditis elegans | Cryo-EM single particle analysis | 3.82 Å | 8P2M |
| AaNGT complexed to UDP and a peptide | Aggregatibacter aphrophilus | X-ray diffraction | 2.8 Å | 8P0Q |
| Insulin receptor-related receptor's transmembrane domain | Homo sapiens | SOLUTION NMR | / | 7PL4 |
| Insulin receptor-related receptor (IRR) in apo-state captured at pH 7 | Homo sapiens | Cryo-EM single particle analysis | 3.3 Å | 7TYJ |
| The kinase domain of RTKC8 | Monosiga brevicollis | X-ray diffraction | 1.95 Å | 8E4T |
| Extracellular WNT-binding module | Drosophila melanogaster | X-ray diffraction | 1.75 Å | 7ME4 |
| The DDR1 Kinase Domain Bound To SR302 | Homo sapiens | X-ray diffraction | 2.3 Å | 7BCM |
| Anaplastic lymphoma kinase (ALK) extracellular ligand binding region 673-1025 | Homo sapiens | SOLUTION NMR | / | 7MZW |
| TG domain of LTK | Homo sapiens | X-ray diffraction | 1.3 Å | 7NX1 |
| Mutation-induced inactivation of Human muscarinic receptor M4 | Homo sapiens | X-ray diffraction | 3 Å | 6KP6 |
| MiniGq-coupled hM3R in complex with iperoxo (local refinement) | Homo sapiens | Cryo-EM single particle analysis | 2.56 Å | 8EA0 |
| MiniGo-coupled hM4Di in complex with DCZ | Homo sapiens | Cryo-EM single particle analysis | 2.7 Å | 8E9X |
| apo state of class C GPCR | Homo sapiens | Cryo-EM single particle analysis | 3.96 Å | 7DGD |
| M1-StaR-T4L in complex with 77-LH-28-1 | Homo sapiens | X-ray diffraction | 2.17 Å | 6ZFZ |
| Histamine receptor H3R in complex with antagonist PF03654746 | Homo sapiens | X-ray diffraction | 2.6 Å | 7F61 |
| Inactive 5-HT5AR in complex with AS2674723 | Homo sapiens | X-ray diffraction | 2.8 Å | 7UM4 |
| OX2R bound to the insomnia drug lemborexant. | Homo sapiens | X-ray diffraction | 2.89 Å | 7XRR |
Table 1. Structural research of novel receptors.
Creative Biostructure provides NMR spectroscopy, X-ray crystallography, and cryo-electron microscopy (cryo-EM) to assist clients in studying novel receptor structures, in order to further understand the disease mechanisms caused by receptor deficiency or to develop new drugs and therapeutic therapies.
We have long been committed to the study of structural biology and membrane proteins. Our experts have extensive experience in determining membrane protein structures. If you are interested in our services, please contact us for more details.
References
- Zarkadas E, et al. Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor. Neuron. 2022. 110(8):1358-1370.
- Miyazawa A, et al. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003. 423(6943):949-955.
- Alexander, et al. Catalytic Receptors. British Journal of Pharmacology. 2007. 150:122-127.
- Wettschureck N, et al. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005. 85(4):1159-1204.