Structural Research of Electron Transport Chain Complex IV
The electron transport chain (ETC) in mitochondria is made up of four complexes that transfer electrons from NADH and FADH2 to oxygen, which causes proton translocation across the inner mitochondrial membrane. Complex IV, also called cytochrome c oxidase (CcO), is the terminal enzyme in the ETC and is responsible for reducing dioxygen to water and coupling the released energy to proton translocation. Bovine CcO (bCcO) is a member of this family and has a molecular weight of about 400 kDa. It is composed of two identical parts, each made up of 13 subunits and four redox centers: a dinuclear copper center (CuA), a heme group (heme a), and a binuclear center composed of another heme (heme a3) and another copper (CuB).
The structural determination of CcO is challenging due to its large size, complex structure, and involvement of redox-active metal centers. Recent advancements in X-ray crystallography and cryo-electron microscopy have enabled high-resolution structural analysis of CcO, providing insights into its mechanism of action. A recent study describes the determination of the structure of bCcO in the CO-bound state at high resolution using serial femtosecond X-ray crystallography (SFX) with an X-ray free electron laser. Furthermore, the study presents an equivalent structure of bCcO at a resolution of 1.95 Å, obtained using a synchrotron light source. In the SFX structure, the CO is coordinated to the heme a3 iron atom, while in the synchrotron structure, the Fe-CO bond is cleaved, and CO relocates to a new site near CuB, resulting in CuB moving closer to the heme a3 iron by approximately 0.38 Å. The ligand binding to the heme a3 iron in the SFX structure triggers an allosteric structural transition, involving partial unwinding of the helix-X between heme a and a3, which establishes a communication linkage between the two heme groups, setting the stage for proton translocation during the ensuing redox chemistry.
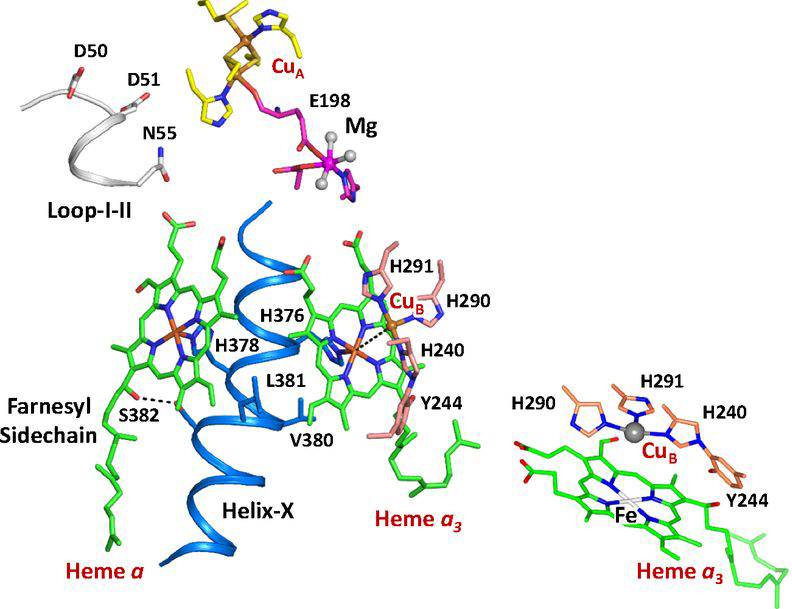 Figure 1. Active site structure of bCcO. (Ishigami I, et al., 2019)
Figure 1. Active site structure of bCcO. (Ishigami I, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Cytochrome C Oxidase, aa3 | Bos taurus | X-ray diffraction | 2.80 Å | 1OCC |
| Fully Oxidized Cytochrome C Oxidase, aa3 | Bos taurus | X-ray diffraction | 1.80 Å | 1V54 |
| Cytochrome C Oxidase, aa3 with bound cyanide | Bos taurus | X-ray diffraction | 2.00 Å | 3X2Q |
| Cytochrome C Oxidase in complex with cytochrome c | Bos taurus | X-ray diffraction | 2.00 Å | 5IY5 |
| Cytochrome C Oxidase at neutral pH | Bos taurus | X-ray diffraction | 1.77 Å | 5XDQ |
| Cytochrome C Oxidase with bound CO by serial femtosecond x-ray | Bos taurus | X-ray diffraction | 2.30 Å | 5W97 |
| Cytochrome c oxidase with bound azide (4-day soak, 20 mM azide) | Bos taurus | X-ray diffraction | 1.85 Å | 5Z84 |
| SFX structure of oxidized cytochrome c oxidase | Bos taurus | X-ray diffraction | 2.90 Å | 6NMP |
| Cytochrome C Oxidase under low-dose x-ray conditions | Bos taurus | X-ray diffraction | 1.90 Å | 6J8M |
| Cytochrome C Oxidase, monomeric, fully-oxidized state | Bos taurus | X-ray diffraction | 1.85 Å | 6JY3 |
| Cytochrome C Oxidase with P-form and F-form intermediates | Bos taurus | X-ray diffraction | 1.80 Å | 6JUW |
| Cytochrome C Oxidase, catalytic intermediate, IO10 | Bos taurus | X-ray diffraction | 1.74 Å | 7D5X |
| Cytochrome C Oxidase, oxidized, with reduced metal centers induced by synchrotron X-ray exposure | Bos taurus | X-ray diffraction | 2.35 Å | 7TIH |
| Cytochrome C Oxidase, fully oxidized state | Bos taurus | X-ray diffraction | 1.50 Å | 5B1A |
| Cytochrome C Oxidase, apo form | Bos taurus | X-ray diffraction | 2.20 Å | 7XMA |
| Cytochrome C Oxidase with bound Ca2+, fully oxidized state (expressed in Bos taurus) | Bos taurus | X-ray diffraction | 1.70 Å | 8H8R |
| Cytochrome C Oxidase, aa3 (expressed in E. coli) | Paracoccus denitrificans | X-ray diffraction | 2.70 Å | 1AR1 |
| Cytochrome C Oxidase, aa3, Fully Oxidized (expressed in E. coli) | Paracoccus denitrificans | X-ray diffraction | 3.00 Å | 1QLE |
| Cytochrome C Oxidase, aa3, N131D variant (expressed in E. coli) | Paracoccus denitrificans | X-ray diffraction | 2.32 Å | 3EHB |
| Cytochrome C Oxidase, aa3 (expressed in E. coli) | Paracoccus denitrificans | X-ray diffraction | 2.25 Å | 3HB3 |
| CBB3 cytochrome oxidase | Stutzerimonas stutzeri | X-ray diffraction | 3.20 Å | 3MK7 |
| Cytochrome ba3 | Thermus thermophilus HB8 | X-ray diffraction | 2.40 Å | 1EHK |
| Cytochrome ba3 with bound xenon (expressed in Thermus thermophilus) | Thermus thermophilus | X-ray diffraction | 3.37 Å | 3BVD |
| caa3-type cytochrome oxidase | Thermus thermophilus HB8 | X-ray diffraction | 2.36 Å | 2YEV |
| Cytochrome C Oxidase wild-type | Cereibacter sphaeroides | X-ray diffraction | 2.30 Å | 1M56 |
| Cytochrome C Oxidase, two-subunit catalytic core (expressed in Cereibacter sphaeroides) | Rhodobacter sphaeroides | X-ray diffraction | 2.00 Å | 2GSM |
| Ubiquinol Oxidase, cytochrome bo3 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.50 Å | 1FFT |
| Ubiquinol Oxidase, cytochrome bo3 (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 2.38 Å | 6WTI |
| Ubiquinol Oxidase, cytochrome bo3 in native membrane | Escherichia coli | Cryo-EM single particle analysis | 2.55 Å | 7CUB |
| Ubiquinol Oxidase, cytochrome bo3, apo form (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.09 Å | 7XMC |
| Cytochrome bd-type oxidase (anisotropy corrected) | Geobacillus thermodenitrificans | X-ray diffraction | 3.05 Å | 5DOQ |
| Cytochrome bd-type oxidase in nanodiscs (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 2.68 Å | 6RKO |
| Cytochrome bd-type oxidase in nanodiscs treated w. specific inhibitor aurachin | Escherichia coli | Cryo-EM single particle analysis | 3.30 Å | 6RX4 |
| Cytochrome bd-II type oxidase with bound aurachin D (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 3.00 Å | 7OSE |
| Cytochrome bd-II type oxidase (expressed in E. coli) | Escherichia coli | Cryo-EM single particle analysis | 2.06 Å | 7OY2 |
| Cytochrome bd-type oxidase (expressed in E. coli) | Mycolicibacterium smegmatis | Cryo-EM single particle analysis | 2.79 Å | 7D5I |
| Heme A synthase (HAS) (expressed in E. coli) | Bacillus subtilis | X-ray diffraction | 2.20 Å | 6A2J |
| Cytochrome aa3-600 menaquinol oxidase | Bacillus subtilis | X-ray diffraction | 3.60 Å | 6KOB |
| Cytochrome C Oxidase (expressed in Saccharomyces cerevisiae) | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 3.87 Å | 7Z10 |
| Cox13 subunit of cytochrome C oxidase (expressed in E. coli) | Saccharomyces cerevisiae | Solution NMR | / | 6ZDB |
Table 1. Structural Research of Electron Transport Chain Complex IV.
Structural analysis are crucial in the study of proteins and other biological macromolecules, and Creative Biostructure is a leading provider of such services. Our team of experts uses a variety of cutting-edge techniques to provide clients with high-quality structural information, such as X-ray crystallography and cryo-electron microscopy (cryo-EM).
Cryo-EM is a powerful structural biology technique that has revolutionized the field of protein structure determination in recent years. It allows the imaging of large and complex macromolecular assemblies, such as membrane proteins and protein complexes, at near-atomic resolution. We offer a range of cryo-EM services, including sample preparation, data acquisition, image processing, and structure determination. In addition to cryo-EM, X-ray crystallography is another commonly used technique for the determination of protein structures. Our team of experts has extensive experience in protein crystallization, data collection, and structure determination using X-ray crystallography. We also offer a range of related services, such as ligand binding studies and protein-protein interaction analysis.
Whether you require cryo-EM or X-ray crystallography services, or a combination of both, Creative Biostructure has the expertise and resources to deliver results that exceed your expectations. Contact us to learn more about our structural analysis services and how we can help advance your research.
References
- Ishigami I, et al. Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature. Proceedings of the National Academy of Sciences. 2017, 114(30): 8011-8016.
- Ishigami I, et al. Snapshot of an oxygen intermediate in the catalytic reaction of cytochrome c oxidase. Proceedings of the National Academy of Sciences. 2019, 116(9): 3572-3577.