RNA isolation and purification are fundamental techniques in molecular biology used to extract high-quality RNA from biological samples for various applications, including gene expression analysis, sequencing, and structural studies. RNA molecules, especially messenger RNA (mRNA), play a critical role in the flow of genetic information within cells, from DNA to protein synthesis. High quality RNA is essential for downstream applications such as reverse transcription PCR (RT-PCR), next-generation sequencing (NGS), and microRNA (miRNA) profiling. However, RNA's inherent instability, susceptibility to degradation by ribonucleases (RNases), and contamination by genomic DNA pose significant challenges.
RNA Structure and Types
RNA is a nucleic acid composed of ribonucleotides linked by phosphodiester bonds. Unlike DNA, RNA contains the sugar ribose, which has a hydroxyl group at the 2' position, making RNA more susceptible to hydrolysis and degradation. The four bases found in RNA are adenine (A), guanine (G), cytosine (C), and uracil (U), with uracil replacing thymine found in DNA. RNA exists in several forms, each with a different function:
- mRNA (messenger RNA): Carries genetic information from DNA to ribosomes, where it is translated into proteins.
- rRNA (ribosomal RNA): Forms the core structural and catalytic components of ribosomes, which are the cellular machines responsible for protein synthesis.
- tRNA (transfer RNA): Transfers specific amino acids to the ribosome during protein synthesis.
- Non-coding RNAs (ncRNAs): Includes microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and small nucleolar RNAs (snoRNAs), which play regulatory and structural roles within the cell.
RNA isolation and purification can be targeted to specific RNA types or can involve the extraction of total RNA, which includes all RNA species present in a given sample.
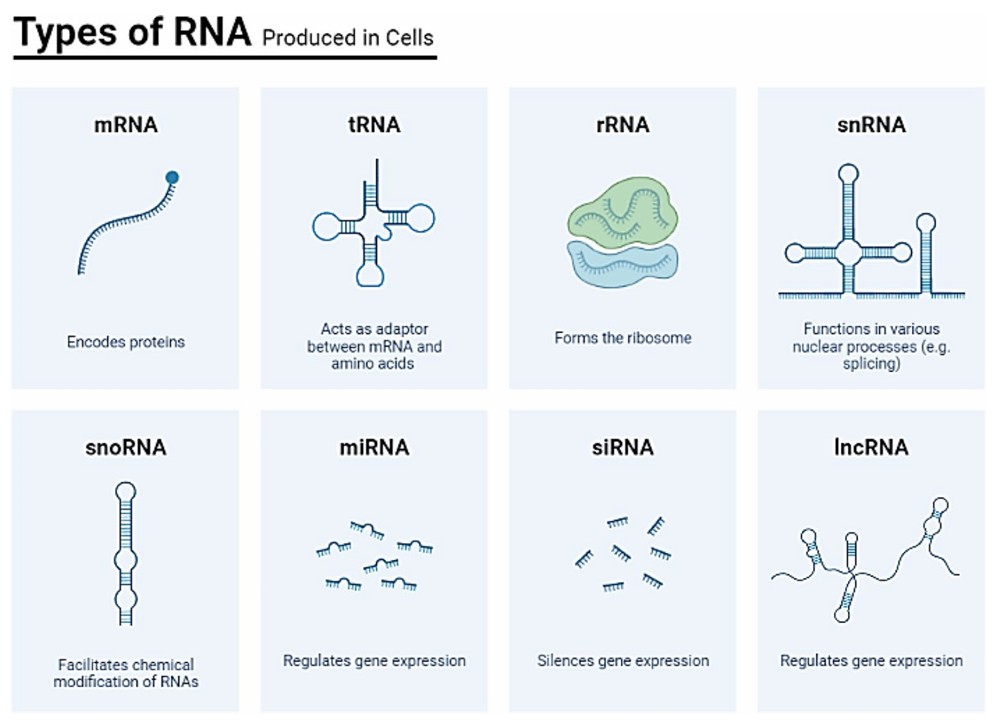 Figure 1. Diversity of RNA types and their functionalities in a cell. It highlights messenger RNA (mRNA) as the template for protein synthesis, transfer RNA (tRNA) responsible for delivering amino acids to the ribosome during translation, ribosomal RNA (rRNA) as a structural component of ribosomes, small nuclear RNA (snRNA) involved in RNA splicing, small nucleolar RNA (snoRNA) participating in ribosomal RNA modification, microRNA (miRNA) regulating gene expression, long non-coding RNA (lncRNA) contributing to diverse cellular processes, small interfering RNA (siRNA) engaging in RNA interference, and other RNA species with specialized functions, providing a comprehensive overview of the functional diversity of cellular RNA molecules. (Suleiman et al., 2024)
Figure 1. Diversity of RNA types and their functionalities in a cell. It highlights messenger RNA (mRNA) as the template for protein synthesis, transfer RNA (tRNA) responsible for delivering amino acids to the ribosome during translation, ribosomal RNA (rRNA) as a structural component of ribosomes, small nuclear RNA (snRNA) involved in RNA splicing, small nucleolar RNA (snoRNA) participating in ribosomal RNA modification, microRNA (miRNA) regulating gene expression, long non-coding RNA (lncRNA) contributing to diverse cellular processes, small interfering RNA (siRNA) engaging in RNA interference, and other RNA species with specialized functions, providing a comprehensive overview of the functional diversity of cellular RNA molecules. (Suleiman et al., 2024)
Challenges in RNA Isolation
RNA Instability and Degradation
RNA is highly labile due to RNases, which are ubiquitous in the environment and in biological samples. Even trace amounts of RNases can degrade RNA within minutes, requiring stringent precautions such as the use of RNase-free reagents and equipment.
Genomic DNA Contamination
Residual genomic DNA in RNA samples can lead to false positives in RT-PCR because DNA can serve as a template during amplification. Studies have shown that no RNA isolation method consistently eliminates DNA contamination, requiring post-isolation treatments such as DNase I digestion.
Complex Sample Matrices
- Plant and fungal samples: High levels of polysaccharides, polyphenols, and secondary metabolites interfere with RNA extraction.
- Bacterial samples: Rigid cell walls and rapid RNA degradation require rapid lysis methods. A 2021 protocol combining formamide-based lysis with chitosan-modified silica purification enables efficient RNA isolation from E. coli in 15 minutes.
RNA Isolation Methodologies
Isolation of RNA from biological samples is typically performed by extraction methods that exploit the differences between RNA and DNA, as well as the physical properties of cellular components. The general principle is to disrupt cells to release RNA, followed by separation of RNA from DNA, proteins, and other cellular debris. The most commonly used techniques for RNA isolation include:
Organic Solvent Extraction (TRIzol Method)
One of the most widely used methods for RNA isolation is the TRIzol reagent (also known as TRI reagent), which is based on a single-step phenol-chloroform extraction process. The process involves:
- Cell Lysis: The biological sample is mixed with TRIzol reagent, which lyses cells and dissociates proteins.
- Phase Separation: Addition of chloroform causes phase separation into three layers: a lower organic phase (containing proteins), an interphase (containing DNA), and an upper aqueous phase (containing RNA).
- RNA Precipitation: The aqueous phase is removed and treated with isopropanol or ethanol to precipitate RNA.
- RNA Purification: The RNA pellet is washed with ethanol and resuspended in RNase-free water.
This method is versatile and can be used to isolate total RNA from a variety of samples, including tissues, cells and biological fluids. However, it requires careful handling to avoid RNase contamination and to ensure efficient RNA recovery.
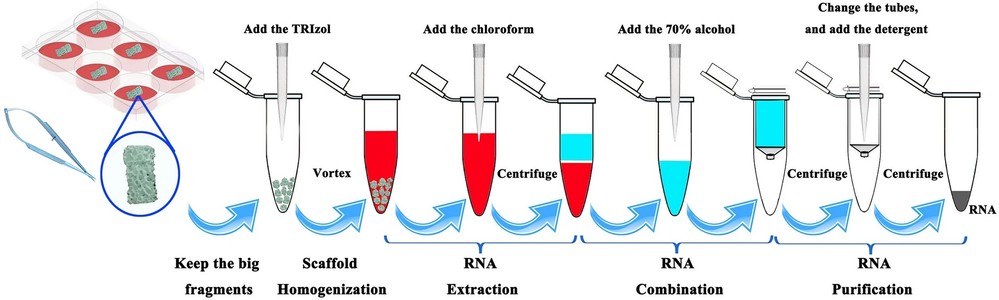 Figure 2. Simplified RNA isolation procedures. (Wang et al., 2022)
Figure 2. Simplified RNA isolation procedures. (Wang et al., 2022)
Silica-Based Column Purification
Silica-based RNA purification methods use spin columns that contain a silica membrane capable of binding RNA in the presence of high concentrations of chaotropic salts. The general procedure involves:
- Sample Lysis: The biological sample is lysed using a buffer that contains chaotropic agents, which disrupt cell membranes and denature proteins.
- RNA Binding: The lysate is applied to a spin column containing a silica membrane. RNA binds to the membrane under high-salt conditions, while contaminants (such as proteins and DNA) are washed away.
- RNA Elution: RNA is eluted from the membrane using low-salt buffer or RNase-free water.
Column-based methods are highly effective for isolating high-quality RNA with minimal contamination. They are commonly used for RNA extraction from cultured cells, tissues, and plasma samples.
Magnetic Bead-Based RNA Isolation
Magnetic bead-based RNA isolation uses paramagnetic beads coated with oligo(dT) or other RNA-specific capture molecules that selectively bind RNA. This method is useful for the isolation of polyadenylated mRNA. The process includes:
- Bead Binding: Magnetic beads coated with oligo(dT) bind to the poly(A) tail of mRNA molecules.
- Separation: The RNA-bead complex is isolated using a magnetic field, and contaminants are washed away.
- RNA Elution: RNA is eluted from the beads using a low-salt buffer or RNase-free water.
Magnetic bead-based methods are highly efficient and can be automated for high-throughput applications. They are often used in RNA-seq and other transcriptomic analyses.
RNA Purification Protocols and Considerations
RNA purification involves several steps to ensure high quality, intact RNA suitable for downstream applications. Several factors need to be considered:
RNase Inhibition
RNases are enzymes that degrade RNA, and they are found in virtually all biological materials. To prevent RNA degradation during isolation, RNase contamination must be avoided. This includes:
- Using RNase-free reagents, tubes, and pipette tips.
- Working in clean, RNase-free environments, such as designated RNA workspaces.
- Adding RNase inhibitors to prevent RNA degradation during the process.
RNA Quality Assessment
The quality of RNA is critical for many downstream applications. Several techniques are used to assess RNA quality:
- Agarose Gel Electrophoresis: RNA samples are run on an agarose gel and visualized under UV light after staining with a dye such as ethidium bromide. High-quality RNA shows distinct 18S and 28S ribosomal RNA (rRNA) bands, while degraded RNA appears as smeared bands.
- Nanodrop Spectrophotometry: The absorbance at 260 nm is measured to estimate the RNA concentration. The 260/280 ratio should be around 2.0, indicating minimal protein contamination.
- Bioanalyzer/Fragment Analyzer: These instruments use capillary electrophoresis to provide a more detailed analysis of RNA integrity, generating an RNA Integrity Number (RIN), which is a standardized measure of RNA quality.
- RT-PCR Efficiency: RNA quality is validated by amplifying housekeeping genes (e.g., GAPDH). Contaminating DNA is detected using "no reverse transcriptase" controls.
- Microarray and Sequencing: High-quality RNA ensures accurate gene expression profiling, as seen in miRNA studies of plant senescence.
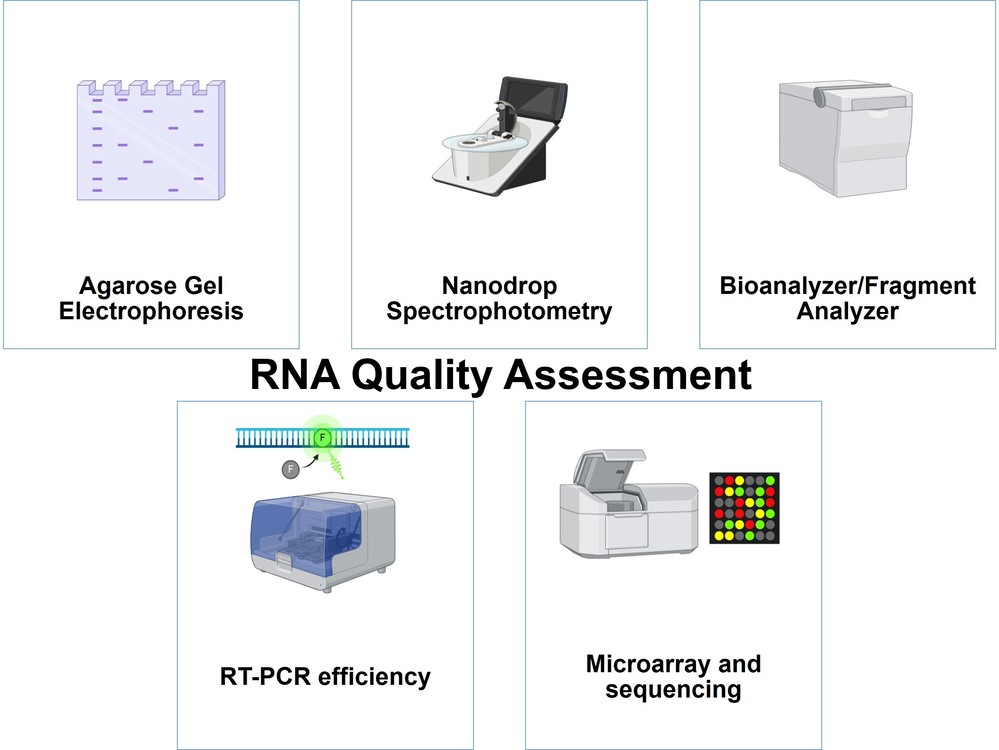 Figure 3. Techniques used for RNA quality assessment.
Figure 3. Techniques used for RNA quality assessment.
Advanced RNA Isolation Technologies
Advancements in RNA isolation have led to the development of more efficient, high-throughput, and automated techniques. Some of these include:
RNA Isolation from Formalin-Fixed, Paraffin-Embedded (FFPE) Samples
RNA extraction from FFPE tissue samples is challenging due to the cross-linking of RNA to proteins and the degradation caused by formalin fixation. Specialized kits and methods, such as the use of protease digestion and optimized buffers, have been developed to release and purify RNA from these samples. These techniques are critical for archival tissue analysis in clinical diagnostics and research.
Single-Cell RNA Isolation
Isolation of RNA from single cells is a rapidly growing field, driven by the need to understand cellular heterogeneity. Single-cell RNA sequencing (scRNA-seq) requires the isolation of RNA from individual cells without contamination from surrounding cells. Techniques such as microfluidic-based isolation, droplet-based methods (e.g. 10x Genomics) and laser capture microdissection are used to extract RNA from single cells or small cell populations.
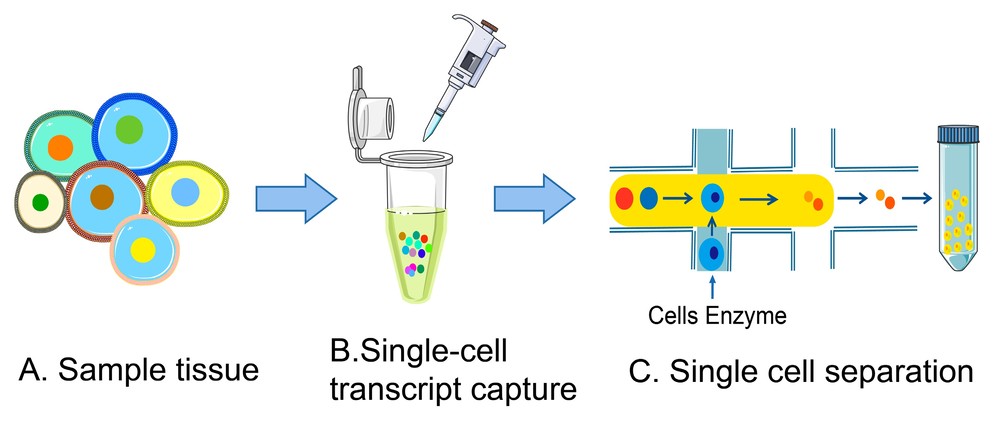 Figure 4. Single-cell RNA isolation procedures. (A) Collect cells from tissue samples. (B) Single-cell capture process. (C) Cell isolation process. (Adapted from Wang et al., 2023)
Figure 4. Single-cell RNA isolation procedures. (A) Collect cells from tissue samples. (B) Single-cell capture process. (C) Cell isolation process. (Adapted from Wang et al., 2023)
High-Throughput Automation
Automated RNA extraction systems can handle large numbers of samples simultaneously, significantly reducing manual labor and variability. These systems use liquid handling robotics, integrated RNA isolation protocols, and quality control features to streamline RNA extraction for large-scale studies, such as transcriptome-wide association studies (TWAS) or large-scale clinical trials.
Applications of RNA Isolation
Gene Expression Analysis
- miRNA profiling: Critical for studying plant senescence, where miRNAs regulate stress responses and developmental transitions. Isolation methods must preserve small RNA species, often achieved through silica-based kits with size-selective binding.
- RT-qPCR: Requires DNA-free RNA to avoid false positives.
Clinical Diagnostics
- Viral RNA detection: Rapid isolation protocols are essential for diagnosing pathogens like SARS-CoV-2. Formamide-based methods offer speed, while chitosan-modified systems enhance purity.
- Cancer biomarkers: High-purity RNA enables the identification of RNA modifications like 2'-O-methylation (Nm), which influence mRNA stability and immune evasion in viruses.
Environmental Monitoring
- Bacterial gene expression: Tracking toxin-induced changes in E. coli using rapid isolation methods supports bioremediation research.
Case Study
Case 1: A Novel Method to Purify Small RNAs from Human Tissues for Methylation Analysis by LC-MS/MS
This study presents a novel two-step method to purify small RNAs (miRNA, piRNA, and tsRNA) from human tissues for LC-MS/MS analysis of their methylation modifications. Methylation of small RNAs is crucial for their biogenesis and function, but LC-MS/MS requires high-purity RNA, which is difficult to obtain due to sequence similarity among small RNAs.
The method consists of two main steps: RNA Pulldown Assay and Enzyme Protection Assay. In the RNA Pulldown Assay, the target small RNA is captured with a biotinylated antisense DNA oligonucleotide, followed by binding to streptavidin magnetic beads and elution with DEPC water. In the enzyme protection assay, the small RNA is paired with complementary single-stranded DNA to form a hybrid duplex, which is then treated sequentially with exonuclease I, nuclease S1, and DNase I to remove non-target sequences. This optimized method successfully purified miR-21-5p, miR-26-5p, piR-020485, and tsRNA from lung and sperm tissues, allowing accurate detection of their 2′-O methylation by LC-MS/MS.
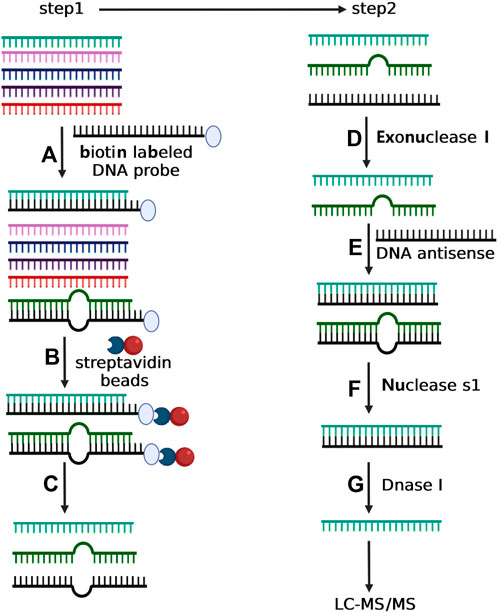 Figure 5. Depiction of two-step experimental strategy for isolating selective small RNA. In step 1, total small RNA was incubated with the biotin-labeled DNA oligonucleotide with complementary sequence to the target small RNA (A). The mixture of total small RNA and biotin-labeled DNA oligonucleotide was then incubated with streptavidin magnetic particle (B) followed by magnetic separation (C). In step 2, the purified small RNA from Step 1 was treated with exonuclease I to degrade the possible residual DNA oligonucleotide (D), and next incubated with a DNA antisense to form RNA/DNA duplex (E). The sample was then incubated with nuclease S1 to eliminate the incomplete complementary small RNAs (F). Finally, the purified small RNA from step f was treated with DNAse I to eliminate the DNA antisense (G). (Yang et al., 2022)
Figure 5. Depiction of two-step experimental strategy for isolating selective small RNA. In step 1, total small RNA was incubated with the biotin-labeled DNA oligonucleotide with complementary sequence to the target small RNA (A). The mixture of total small RNA and biotin-labeled DNA oligonucleotide was then incubated with streptavidin magnetic particle (B) followed by magnetic separation (C). In step 2, the purified small RNA from Step 1 was treated with exonuclease I to degrade the possible residual DNA oligonucleotide (D), and next incubated with a DNA antisense to form RNA/DNA duplex (E). The sample was then incubated with nuclease S1 to eliminate the incomplete complementary small RNAs (F). Finally, the purified small RNA from step f was treated with DNAse I to eliminate the DNA antisense (G). (Yang et al., 2022)
Case 2: Serum Insights: Leveraging the Power of miRNA Profiling as an Early Diagnostic Tool for Non-Small Cell Lung Cancer
This study investigates the potential of serum miRNAs as non-invasive biomarkers for early-stage non-small cell lung cancer (NSCLC). Using next-generation sequencing, the researchers profiled miRNAs from 71 NSCLC patients and 47 individuals with non-cancerous lung conditions. They identified 28 upregulated miRNAs in NSCLC that were associated with key cancer-related pathways.
To improve diagnostic accuracy, a gradient boosting decision tree classifier was developed using 2588 miRNAs. The model achieved high accuracy, sensitivity and specificity.
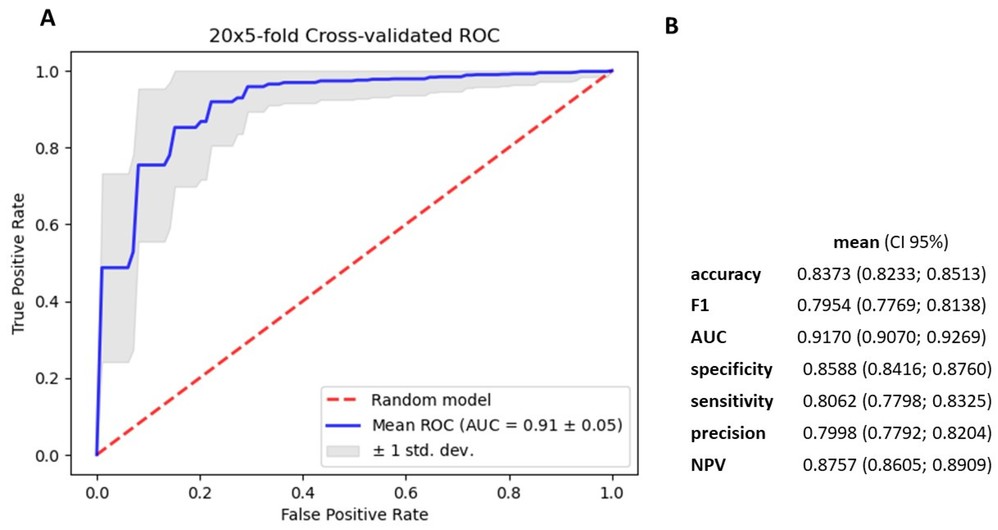 Figure 6. Metrics for gradient boosting decision tree model based on 2588 miRNA. (A). mean ROC ± SD curve, and mean AUC for classifier (B). mean and 95% CI of accuracy, f1–score metrics, AUC, specificity, sensitivity, precision and NPV for differentiating cancerous and noncancerous patients' serum. (Charkiewicz et al., 2023)
Figure 6. Metrics for gradient boosting decision tree model based on 2588 miRNA. (A). mean ROC ± SD curve, and mean AUC for classifier (B). mean and 95% CI of accuracy, f1–score metrics, AUC, specificity, sensitivity, precision and NPV for differentiating cancerous and noncancerous patients' serum. (Charkiewicz et al., 2023)
Creative Biostructure is a leading expert in structural biology, offering a comprehensive range of RNA-related services. Our expertise includes RNA structure characterization, analysis of RNA structure and dynamics, and characterization of protein-RNA interactions,among others. We provide high-quality, reliable solutions tailored to your research needs. Contact us today to discuss your project and learn more about our specialized RNA analysis services!
References
- Charkiewicz R, Sulewska A, Mroz R, et al. Serum insights: leveraging the power of miRNA profiling as an early diagnostic tool for non-small cell lung cancer. Cancers. 2023;15(20):4910. doi:10.3390/cancers15204910
- Hu P, Zhang W, Xin H, Deng G. Single cell isolation and analysis. Front Cell Dev Biol. 2016;4. doi:10.3389/fcell.2016.00116
- Suleiman AA, Al-Chalabi R, Shaban SA. Integrative role of small non-coding RNAs in viral immune response: a systematic review. Mol Biol Rep. 2024;51(1):107. doi:10.1007/s11033-023-09141-6
- Yang R, Li J, Wu Y, et al. A novel method to purify small RNAs from human tissues for methylation analysis by LC-MS/MS. Front Mol Biosci. 2022;9:949181. doi:10.3389/fmolb.2022.949181
- Wang Q, Wang W, Zhang P, et al. A simplified method for RNA isolation from biofabricating hydroxyapatite scaffolds and identification of appropriate reference genes. J Med Biol Eng. 2022;42(5):713-721. doi:10.1007/s40846-022-00744-1
- Wang S, Sun ST, Zhang XY, et al. The evolution of single-cell RNA sequencing technology and application: progress and perspectives. International Journal of Molecular Sciences. 2023;24(3):2943. doi:10.3390/ijms24032943