Structural Research of Orthomyxoviridae
The Orthomyxoviridae includes enveloped viruses with negative-sense, single-stranded, and segmented RNA genomes. The family consists of seven genera that infect vertebrates and spread influenza epidemics around the world each year. Orthomyxoviruses (influenza viruses) are classified into four types based on antigenic differences in nucleoprotein (NP) and matrix (M) proteins: types A, B, C, and D. Of these, influenza A viruses (IAV) are responsible for recurrent infections and occasionally devastating epidemics in humans and animals.
Influenza is usually controlled by vaccines around the world, but new strains of influenza viruses are constantly emerging, resulting in limited effectiveness of vaccination. Therefore, new preventive and therapeutic approaches are needed. Knowledge of the structure of influenza viruses and their interactions with the hosts is essential for a better understanding of the mechanisms of viral infection and replication, leading to the proposal of more effective antiviral approaches.
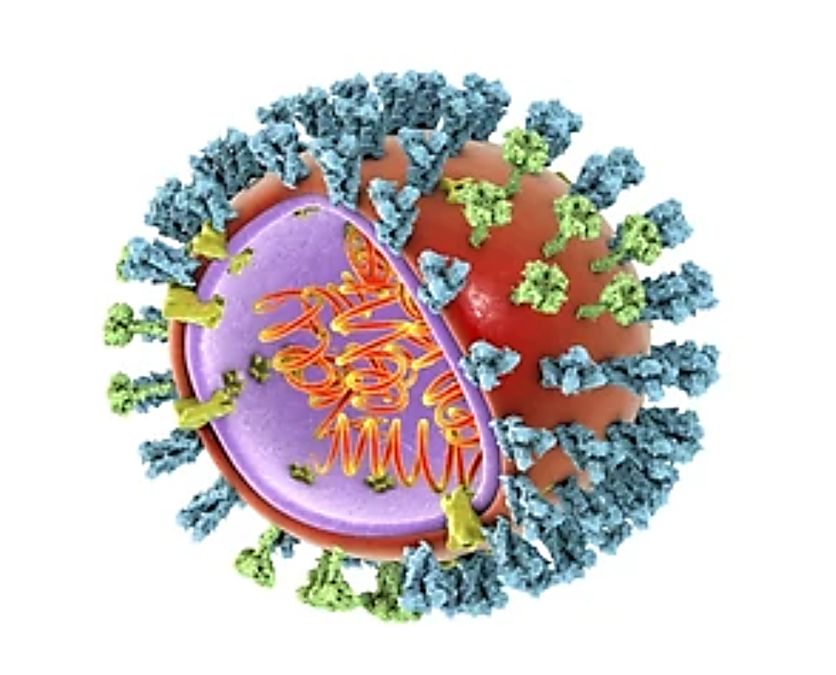
Structural Features of Influenza Viruses
There are two types of influenza viruses, filamentous and polymorphic, most of which are polymorphic and often have round particles of 70 - 120 nm when stained negatively. Unlike other viruses, influenza viruses have approximately equidistantly spaced projections on their surfaces. One is a rod-shaped projection about 10 - 14 nm long and 4 nm wide, triangular in cross-section, containing haemagglutinin, and the other is a cylindrical projection about 9 nm long and 5 nm wide containing neuraminidase. Below the protrusions is a lipid periplasm that surrounds the matrix and nucleic acid proteins.
Structural Research on IAV
The structure of IAV is similar to that of other influenza viruses, with a diameter of about 80 to 120 nm. It is usually roughly spherical, but filamentous influenza viruses also exist. To date, several structures of the IAV ribonucleoprotein (RNP) complex have been resolved by cryo-electron microscopy (cryo-EM) at ~20 Å resolution. Unlike other viruses, the genome of IAV contains eight single-stranded negative-sense RNA fragments, which are complexed with nuclear proteins to form a helically symmetric nucleocapsid encoding a total of 11 proteins. The most representative of these proteins are haemagglutinin and neuraminidase, which are recognized by antibodies and are targeted by antiviral drugs.
Influenza viruses are a serious public health problem due to the widespread pandemics, and appropriate antiviral measures must be put in place. As a leader in viral structural biology. Creative Biostructure provides customized viral structure analysis services and high-quality virus-like particles (VLPs) products to help clients develop influenza virus vaccines, antiviral drugs, and related prevention and treatment strategies.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V501 | A/BrevigMission/1/ 1918 (H1N1) VLP (BlaM1; HA; NA Proteins) | H1N1 | Mammalian cell recombinant | BlaM1; HA; NA |
| CBS-V508 | A/duck/Korea/A76/2010 (H7N7) VLP (HA Proteins) | H7N7 | Insect cell recombinant | HA |
| CBS-V516 | A/Indonesia/05/2005 (H5N1) VLP (HA Proteins) | H5N1 | Plant recombinant | HA |
| CBS-V532 | A/Taiwan/S02076/2013 (H7N9) VLP (HA; NA; M1 Proteins) | H7N9 | Insect cell recombinant | HA; NA; M1 |
| Explore All Orthomyxoviridae Virus-like Particle Products | ||||
Creative Biostructure is dedicated to the field of viral structural analysis, providing cutting-edge cryo-electron microscopy (cryo-EM) technology, comprehensive structural biology services, and high-quality virus-like particles (VLPs) products. We can delve into the complex structure of viruses and provide valuable insights into the molecular mechanisms, driving groundbreaking discoveries in virology for clients and the development of effective antiviral interventions. Please feel free to contact us to discuss your project.
References
- Rosário-Ferreira N, et al. The Central Role of Non-Structural Protein 1 (NS1) in Influenza Biology and Infection. Int J Mol Sci. 2020. 21(4): 1511.
- Ferhadian D, et al. Structural and Functional Motifs in Influenza Virus RNAs. Front Microbiol. 2018. 9: 559.