Structural Research of PNPT Superfamily
The PNPT superfamily includes the bacterial and eukaryotic cell integrase families, which are involved in the synthesis of cell envelope polymers and protein N-linked glycosylation. MraY (phospho-MurNAc-pentapeptide translocase) is the most extensively researched member of the PNPT superfamily. Many natural nucleoside inhibitors with antimicrobial activity target MraY, and it is therefore considered promising for the development of new antibiotics. In recent years, researchers have analyzed the structure of MraY to promote its clinical use.
Analysis of the function and action mechanism of MraY
MraY catalyzes the transfer of phospho-MurNAc-pentapeptide from UDP-MurNAc-pentapeptide (UM5A) to the lipid carrier undecaprenyl phosphate (C 55 -P) to produce lipid I. It is blocked by five classes of natural nucleoside antibiotics (such as muromycin and clindamycin) and the lytic protein E of phage phiX174, with multiple modes of inhibition. Recently, MraY-targeted natural products have been of interest because of their in vivo efficacy against pathogenic bacteria such as M. tuberculosis, methicillin-resistant S. aureus, and vancomycin-resistant Enterococcus.
Advances in structural research on MraY
Researchers used selenomethionine substituted MraY crystals for single anomalous dispersion phasing and determined the MraY crystal structure at 3.3 Å resolution. MraY is a dimer in the asymmetric unit. The center of the dimer interface is an elliptical tunnel surrounded by hydrophobic amino acids to accommodate the lipid. Each protomer contains 10 transmembrane helices (TM1 to TM10), an interfacial helix (IH), a periplasmic β-hairpin (PB), a periplasmic helix (PH), and five cytoplasmic loops (loop A to loop E). TM9 splits into two helical fragments (TM9a and TM9b). TM9b, together with TM3, TM4, and TM8, surrounds TM5 and creates a cleft around the inner leaflet membrane region of TM8, which serves as the active site.
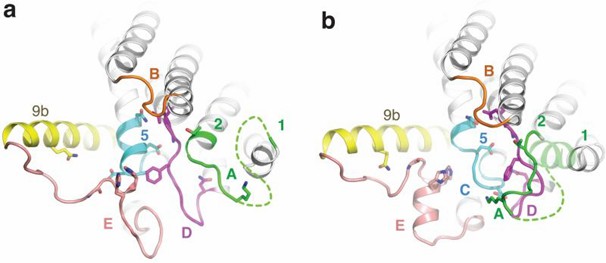 Figure 1. Structure of apoMraY from the cytoplasmic side (a) and bound to the inhibitor (b). (Chung BC, et al., 2016)
Figure 1. Structure of apoMraY from the cytoplasmic side (a) and bound to the inhibitor (b). (Chung BC, et al., 2016)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase | Aquifex aeolicus | X-ray diffraction | 3.3 Å | 4J72 |
| MraY in complex with Muraymycin D2 | Aquifex aeolicus VF5 | X-ray diffraction | 2.95 Å | 5CKR |
| MraY tunicamycin complex | Enterocloster bolteae 90A9 | X-ray diffraction | 2.6 Å | 5JNQ |
| MraY bound to carbacaprazamycin | Aquifex aeolicus VF5 | X-ray diffraction | 2.95 Å | 6OYH |
| MraY bound to capuramycin | Aquifex aeolicus VF5 | X-ray diffraction | 3.62 Å | 6OYZ |
| MraY bound to 3'-hydroxymureidomycin A | Aquifex aeolicus VF5 | X-ray diffraction | 3.7 Å | 6OZ6 |
| MraY bound to a sphaerimicin analogue | Aquifex aeolicus | X-ray diffraction | 3.65 Å | 8CXR |
Table 1. Structural research of PNPT superfamily.
Protein structure analysis is essential for understanding the function of membrane proteins such as the PNPT superfamily. At Creative Biostructure, we offer a range of services for the structural analysis of biomolecules, including X-ray crystallography, cryo-electron microscopy (cryo-EM) and NMR spectroscopy. Our experienced researchers determine the 3D structure of proteins at high resolution, providing clients with accurate data and information.
If you are interested in exploring the structure of the PNPT superfamily or other biomolecules and would like to learn more about the services we offer, please feel free to contact us. Our team is always available to discuss your research needs and provide the best solution for your project.
References
- Chung BC, et al. Structural insights into inhibition of lipid I production in bacterial cell wall synthesis. Nature. 2016.533(7604):557-560.
- Chung BC, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013.341(6149):1012-1016.
- Hering J, et al. Structural basis for selective inhibition of antibacterial target MraY, a membrane-bound enzyme involved in peptidoglycan synthesis. Drug Discov Today. 2018.23(7):1426-1435.