Native Protein Isolation
Native protein isolation is required for various experimental applications such as in vitro biochemical assays and structural studies. Both the amount and purity of the native protein must be sufficient for downstream analysis. Successful native protein isolation is able to preserve the native activity and three-dimensional structure of a target protein during harvest.
Chromatography is the dominant technique for the isolation of native proteins. By exploiting the unique properties of proteins, such as their size, charge, or specific affinity for certain ligands, chromatography provides an efficient and versatile method for purifying proteins from complex biological mixtures. This approach allows for high-resolution separation, ensuring that the target protein can be isolated with high purity and yield while maintaining its functional state.
At Creative Biostructure, we specialize in providing high-quality native protein isolation services to advance your research and development projects. Whether you're studying protein structure, conducting functional assays, or exploring new therapeutic targets, our expert chromatography services ensure that your proteins are isolated in their most active, native form.
Four Basic Steps of Native Protein Isolation
The process of native protein isolation generally involves four essential steps, each of which plays a critical role in maintaining the protein's integrity and ensuring that the final product is suitable for further analysis.
Cell Lysis
The first step in protein isolation is the disruption of the cell membrane to release the intracellular contents, including the protein of interest. This step is crucial, as cells are typically surrounded by membranes that must be broken to access the proteins inside. Various methods of lysis can be used, including mechanical disruption (e.g., homogenization, sonication), chemical lysis (e.g., detergents), and enzymatic lysis (e.g., lysozyme for bacterial cells). The choice of lysis method is determined by the type of sample being processed and the properties of the target protein.
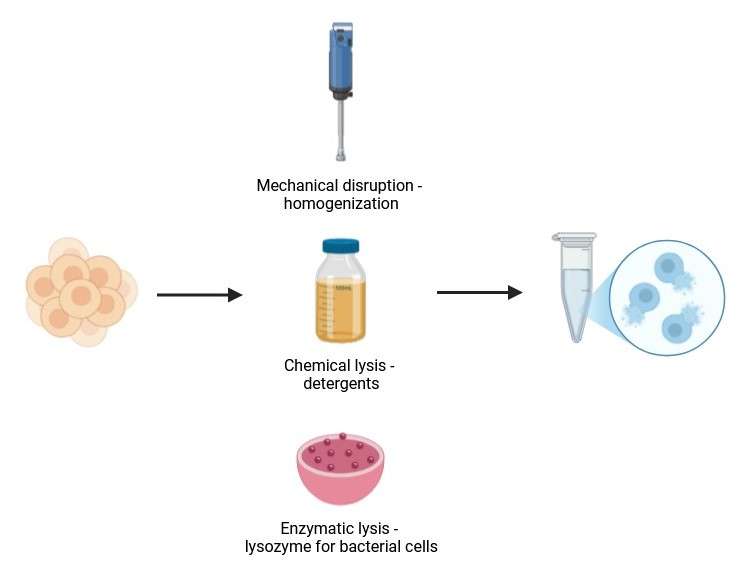 Figure 1. First step of native protein isolation—cell Lysis. (Created with BioRender.com)
Figure 1. First step of native protein isolation—cell Lysis. (Created with BioRender.com)
Protein Binding to a Matrix
After cell lysis, the next step is to capture the proteins on a suitable matrix that can facilitate their subsequent purification. This is often achieved by chromatographic techniques in which proteins bind to a resin or column matrix based on specific characteristics such as charge, size, or affinity for a ligand. Binding is typically performed under mild conditions to preserve the native state and activity of the protein. In affinity chromatography, for example, a specific ligand (such as a metal ion or antibody) is used to selectively capture the target protein.
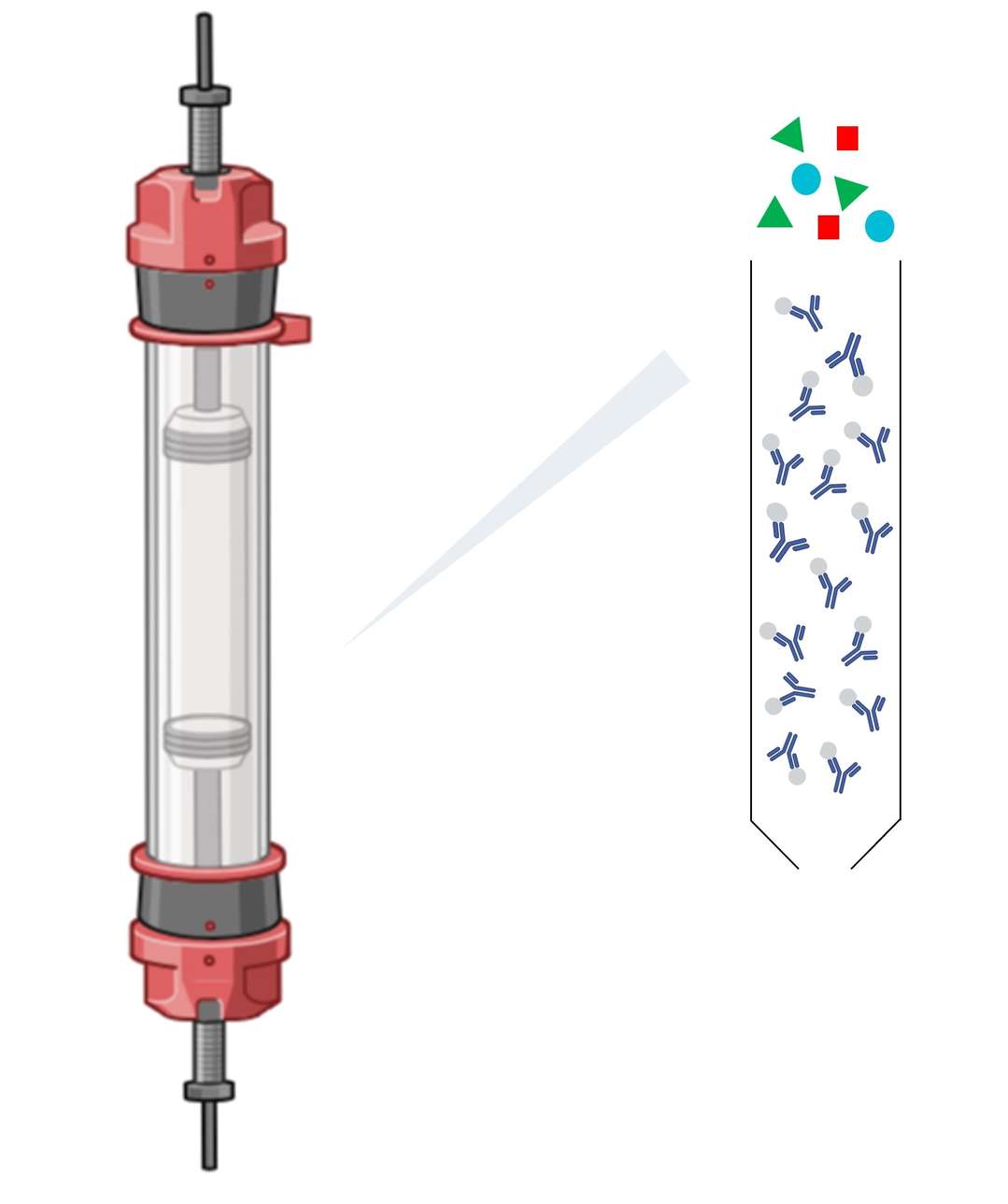 Figure 2. Second step of native protein isolation—protein binding to chromatography column matrix. (Created with BioRender.com)
Figure 2. Second step of native protein isolation—protein binding to chromatography column matrix. (Created with BioRender.com)
Washing
Once the proteins have bound to the matrix, unbound contaminants, including non-target proteins, lipids, and nucleic acids, must be washed away. This step ensures that only the target protein remains attached to the matrix, improving the purity of the isolated protein. The washing conditions—such as buffer composition, pH, and salt concentration—are carefully optimized to remove impurities without disrupting the protein's binding to the matrix or causing denaturation.
Elution
The final step in the isolation process involves eluting the target protein from the matrix. Elution is typically achieved by altering the conditions in the column, such as changing the pH, ionic strength, or adding a competing ligand that displaces the bound protein. In affinity chromatography, for example, imidazole might be used to elute histidine-tagged proteins from a nickel column. The goal is to recover the protein in its native state, ready for further analysis or functional assays.
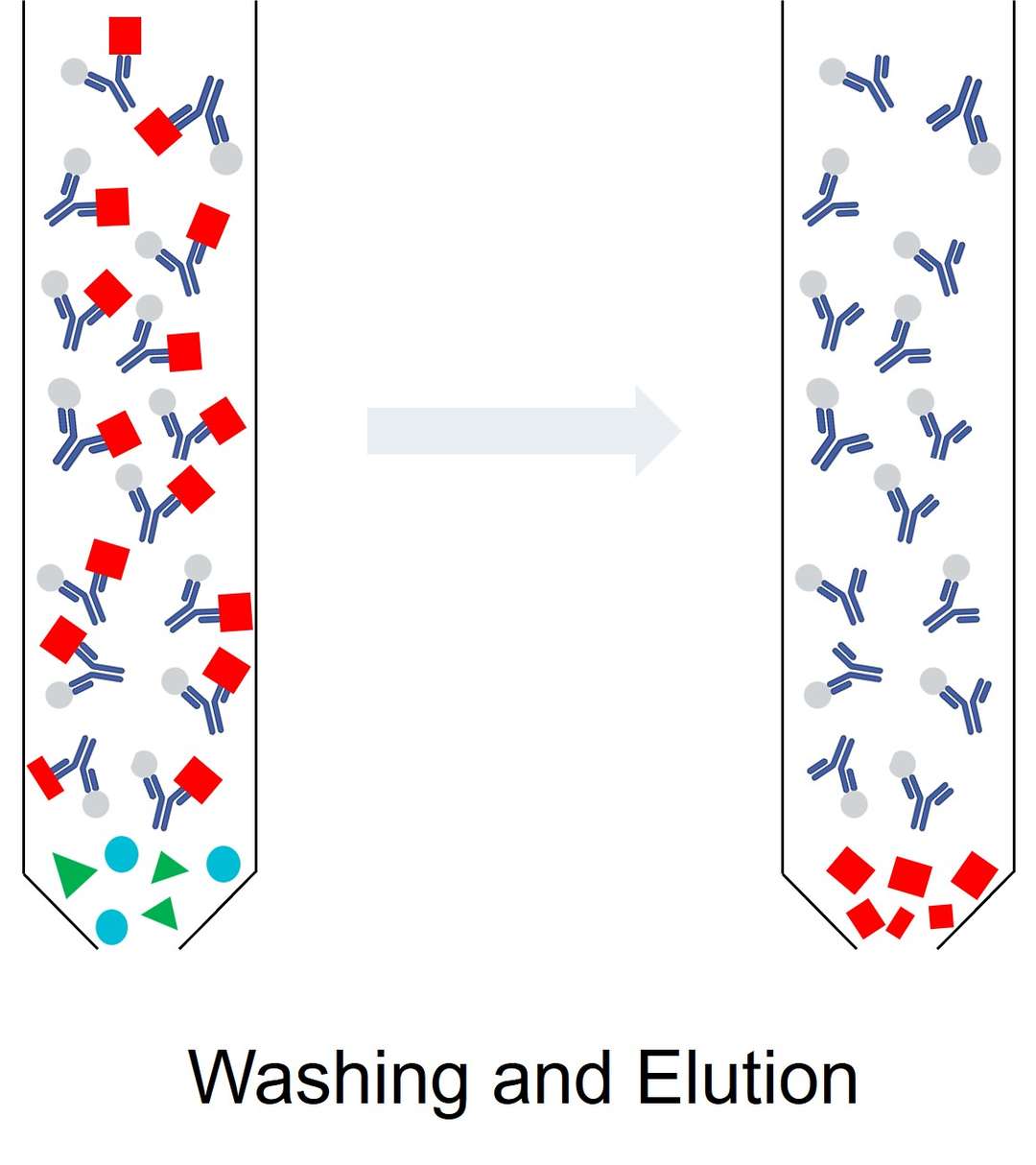 Figure 3. The third and final step of native protein isolation—washing and elution.
Figure 3. The third and final step of native protein isolation—washing and elution.
Common Techniques for Native Protein Isolation
Several chromatography-based techniques are commonly used for native protein isolation, each designed to exploit different protein characteristics. These techniques are often used in combination to achieve high levels of purity and yield.
Affinity Chromatography
Affinity chromatography is one of the most powerful and selective methods for the isolation of native proteins. It is based on the specific binding interaction between the protein and a ligand immobilized on a solid matrix. Common examples include His-tag affinity chromatography (His-tagged Proteins), where a protein with a polyhistidine tag binds to a nickel or cobalt ion on the resin, or antibody-based affinity chromatography, where proteins with specific epitopes bind to antibodies immobilized on the matrix. This method allows highly specific isolation of a target protein with minimal contamination, making it ideal for obtaining pure native proteins.
Ion-Exchange Chromatography
Ion-exchange chromatography separates proteins based on their charge. In this technique, proteins are passed through a column containing a charged resin that attracts proteins of opposite charge. The protein solution is then passed through the column under controlled conditions, and proteins are eluted by changing the salt concentration or pH. This method is particularly useful for isolating proteins with different charge characteristics and is often used in combination with other techniques to achieve a higher degree of purity.
Gel Filtration Chromatography (Size-Exclusion Chromatography)
Gel filtration chromatography (size-exclusion chromatography) separates proteins based on their size. In this technique, proteins are applied to a column filled with porous beads. Larger proteins elute first because they are unable to penetrate the pores of the beads and therefore move more quickly through the column. Smaller proteins spend more time inside the beads and elute later. Gel filtration is often used for final polishing of protein samples, providing a clean separation of proteins based on their size and providing insight into their oligomeric state or quaternary structure.
Reverse-Phase Chromatography
Reverse-phase chromatography is a technique based on hydrophobic interactions in which proteins are separated according to their hydrophobicity. The stationary phase consists of a hydrophobic resin and proteins are eluted based on their affinity for the stationary phase. This technique is particularly useful for separating proteins that differ in their hydrophobic properties and is often used in the purification of membrane proteins or proteins with significant hydrophobic regions.
Troubleshooting
| Problem | Cause | Solution |
| After elution, there is no recombinant protein recovery. | The loading amount of sample is not enough. | Increase the amount of sample loaded or lysate used |
| Protein degraded. | Perform all the native protein isolation steps at 4°C | |
| Make sure that the His-tag is not cleaved during native protein isolation | ||
| Add protease inhibitors during cell/tissue lysis. | ||
| The affinity between recombinant protein and resin is too high. | Increase stringency of elution by decreasing the pH | |
| Increase the imidazole concentration | ||
| Use EDTA or EGTA (10– 100 mM) to strip resin of nickel ions and elute the protein in order to preserve activity | ||
| Native protein was washed out by too stringent washing. | Wash less extensively with wash buffer at an increased pH | |
| Expression level is too low. | Optimize your expression protocols follows the guidelines | |
| There is no protein bonded because of the protein folding. | Try denaturing conditions | |
| Good recombinant- protein recovery but with non-recombinant proteins contamination. | Wash conditions are not sufficiently stringent. | Wash more extensively with wash buffer at a decreased pH |
| Recombinant protein has low affinity for resin; comes off in wash with many contaminating proteins. | Try denaturing conditions | |
| Perform an imidazole gradient elution | ||
| Perform a decreasing pH gradient | ||
| Sample contain other His-rich proteins. | Before perform elution, try an additional high stringency wash at a lower pH (for example, between pH 4 and pH 6) | |
| Further purification of the eluate on a new column after protein dialysis of the eluate against the binding buffer and proper equilibrate the column with binding buffer | ||
| Use another type of column to perform the native protein isolation one more time | ||
| Native protein was washed out by too stringent washing. | Expression levels is too low. | Before perform elution, try an additional high stringency wash at a lower pH (for example, between pH 4 and pH 6) |
| Recombinant protein do not bind to resin tightly. | Try denaturing conditions | |
| Try reverse-chromatography which only works for native protein isolation | ||
| Column become white. | Buffers with Chelating agents which strip the nickel ions from the column. | The column need to recharge thoroughly |
| Column become reddish brown. | Buffers with DTT. | Use β-ME as a reducing agent |
| Detect recombinant protein in the flow through and wash fractions. | Protein overload. | Reduce the amount of protein loaded on the column or increase the amount of resin for native protein isolation |
| Protein precipitation occurs during binding. | Temperature is too low. | Perform native protein isolation at room temperature. |
| Protein aggregation occurs. | You may add solubilization reagents such as 0.1% Tween-20 or 0.1% Triton X-100 or stabilizers such as Mg2+. All buffers may need to add solubilization reagents to maintain protein solubility. | |
| Run column in drip mode in order to prevent protein from dropping out of solution. |
To learn more about how Creative Biostructure's native protein isolation services can support your research or to get started on your next project, contact us today. Our team is ready to provide you with expert guidance and ensure that your protein isolation process is efficient, reliable, and tailored to your exact needs.
