Structural Research of UbiA Prenyltransferases
The UbiA superfamily is a group of intramembrane prenyltransferases that catalyze Mg2+-dependent transfer of hydrophobic polyisoprenoid chains to various receptor molecules. Additionally, UbiA is involved in the synthesis of essential lipophilic compounds in biological membranes, including various quinones, heme, chlorophyll, vitamin E, and structural lipids. The prenyltransferases that produce these compounds are involved in essential physiological processes and human diseases.
Catalytic mechanism of UbiA
Most UbiA superfamily enzymes catalyze the isoprenylation of aromatic substrates to produce the basic skeleton of a variety of lipophilic compounds. The addition of an aliphatic tail confers lipid solubility to these compounds. UbiA catalysis begins with the cleavage of the diphosphate group on the substrate isoprenyl diphosphate (XPP). The joining of the pyrophosphate group of XPP by two aspartic acid-rich motifs in UbiA produces a highly reactive carbon cationic intermediate at the end of the isoprenyl chain. To complete the isoprenylation reaction, the carbon cation undergoes a regiospecific reaction at the interstitial site of the aromatic PHB substrate to form a C-C or C-O bond.
Structural analysis of DGGGPase, a member of the UbiA superfamily
DGGGP synthase (DGGGPase) is a member of the UbiA superfamily that produces membrane core lipids in archaea. Researchers crystallized the purified DGGGPase protein using lipid cubic phase (LCP) and obtained crystals suitable for x-ray diffraction in the presence of DDM detergent. The structure shows that DGGGPase contains nine TM helices forming a flat transmembrane structural domain (TMD) with a large central cavity. The cavity covered by the cytoplasmic structural domain (CPD) consists of three loop/helix regions (CHL23, CHL45, and CHL67) connecting TM2 and TM3, TM4 and TM5, and TM6 and TM7, respectively. Close to the central cavity are two hydrophobic tunnels (one consisting of TM2, TM4, and TM5, and the other of TM6 and TM9) buried in the membrane.
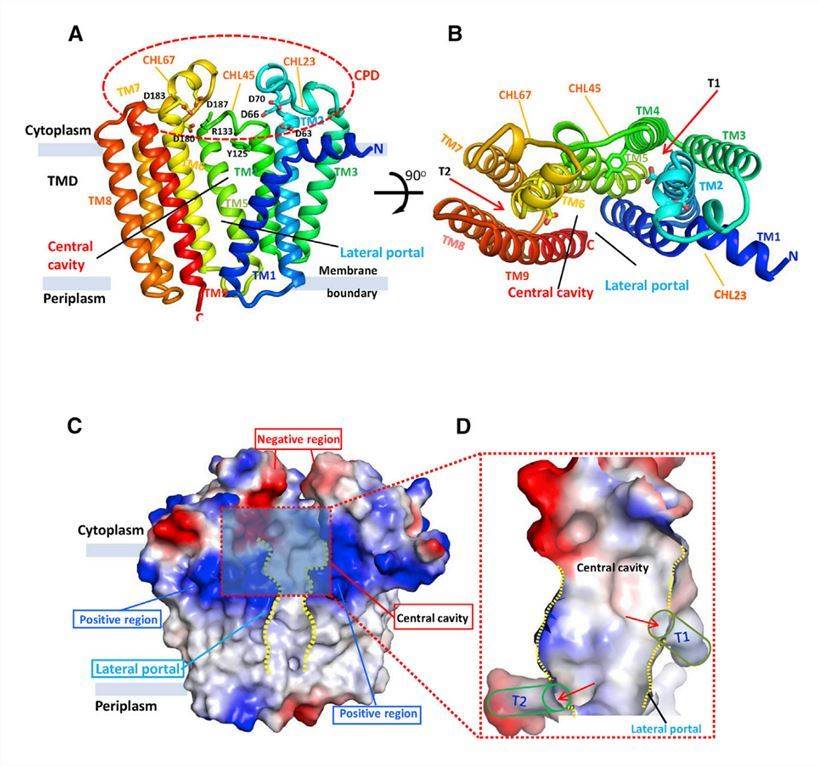 Figure 1. Overall apo-structure of DGGGPase. (Ren S, et al., 2020)
Figure 1. Overall apo-structure of DGGGPase. (Ren S, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Substrate-bound of a UbiA homolog | Aeropyrum pernix K1 | X-ray diffraction | 3.557 Å | 4OD5 |
| UbiA homolog bound to DMAPP and Mg2+ | Archaeoglobus fulgidus DSM 4304 | X-ray diffraction | 2.5025 Å | 4TQ4 |
| Archaeal Lipid Synthase | Methanocaldococcus jannaschii DSM 2661 | X-ray diffraction | 2.3 Å | 6M31 |
| Archaeal synthase | Methanocaldococcus jannaschii DSM 2661 | X-ray diffraction | 2.9 Å | 6M34 |
| UbiA homolog | Archaeoglobus fulgidus DSM 4304 | X-ray diffraction | 3.2023 Å | 4TQ5 |
| UbiA homolog bound to GPP and Mg2+ | Archaeoglobus fulgidus DSM 4304 | X-ray diffraction | 2.4076 Å | 4TQ3 |
| Apo structure of a UbiA homolog | Aeropyrum pernix K1 | X-ray diffraction | 3.301 Å | 4OD4 |
| UbiA homolog bound to Cd2+ | Archaeoglobus fulgidus DSM 4304 | X-ray diffraction | 3.0678 Å | 4TQ6 |
| HMGCR-UBIAD1 Complex State 1 | Cricetulus griseus | Cryo-EM single particle analysis | 3.23 Å | 8DJM |
| HMGCR-UBIAD1 Complex State 2 | Cricetulus griseus | Cryo-EM single particle analysis | 3.33 Å | 8DJK |
| Digeranylgeranylglyceryl phosphate synthase in membranes | Methanocaldococcus jannaschii DSM 2661 | X-ray diffraction | 3.32 Å | 7BPU |
Table 1. Structural research of the UbiA prenyltransferases.
Creative Biostructure is dedicated to protein structure research. Our experts have extensive experience in membrane protein structure determination. We provide X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy platforms to probe protein structures. In addition, we help clients to deeply explore the catalytic mechanism of UbiA prenyltransferases involved in the synthesis of lipophilic compounds.
In addition to membrane protein structure determination, we can accurately analyze other biomolecules including, but not limited to, ribosomes, nucleic acids, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us for a professional solution.
References
- Ren S, et al. Structural and Functional Insights into an Archaeal Lipid Synthase. Cell Rep. 2020. 33(3): 108294.
- Li W. Bringing Bioactive Compounds into Membranes: The UbiA Superfamily of Intramembrane Aromatic Prenyltransferases. Trends Biochem Sci. 2016. 41(4): 356-370.
- Yang Y, et al. Methods for Structural and Functional Analyses of Intramembrane Prenyltransferases in the UbiA Superfamily. Methods Enzymol. 2017. 584: 309-347.