Structural Research of Oxidoreductases
Oxidoreductase is an enzyme that promotes redox reactions, involving the transfer of electrons from one molecule to another. These enzymes play a crucial role in various metabolic pathways, such as cellular respiration, photosynthesis, and detoxification of foreign bodies. Some well-known oxidoreductases include alcohol dehydrogenase, cytochrome P450, and superoxide dismutase.
X-ray crystallography and low-temperature electron microscopy have been used to study the structure of oxidoreductase. The structural information obtained from experiments can help scientists gain insight into the oxidation mechanisms in living organisms.
For example, electron transfer flavoprotein menadione oxidoreductase ABCX (EtfABCX) is a flavin-based complex. In an article published in 2020, cryoelectron microscopy helped scientists clarify the structure and catalytic mechanism of Fix/EtfABCX. In the experiment, scientists used a resolution of 2.9 Å to characterize the structure of membrane-related flavin-based electric bifurcation (FBEB) complex EtfABCX from thermophilic bacteria. The two membrane-related EtfC at the interface between EtfABCX and the dimer forms a super dimer containing two bound quinones. This structure indicates that the low potential electrons bifurcating from EtfAB are most likely to be directly transferred to ferritin, while the high likely electrons reduce quinones through two [4Fe-4S] clusters in EtfX.
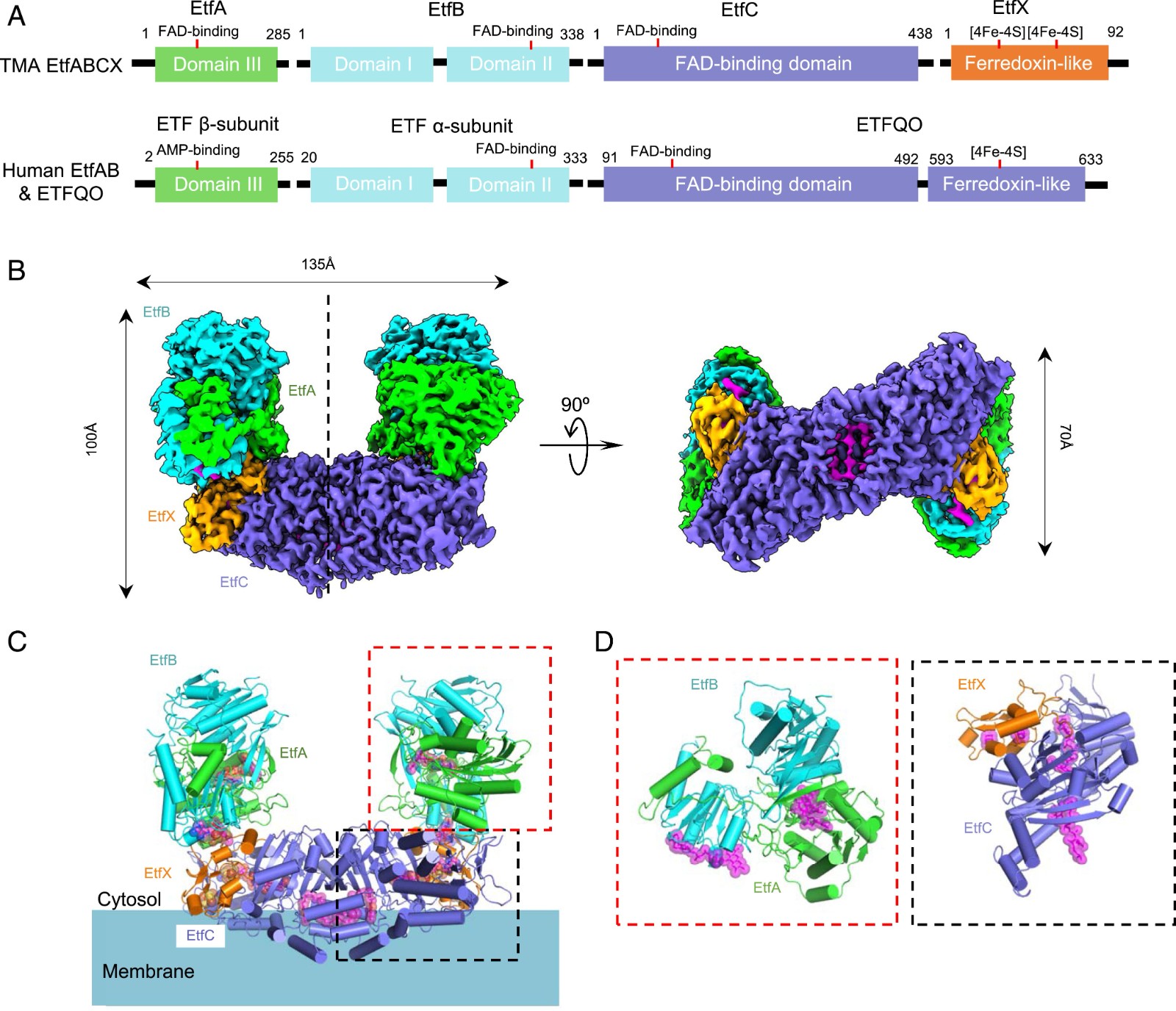 Figure 1. Superdimeric architecture of the Tma EtfABCX. (FENG, et al., 2020)
Figure 1. Superdimeric architecture of the Tma EtfABCX. (FENG, et al., 2020)
The first step of hydrogen sulfide (H2S) metabolism in mitochondria is catalyzed by a whole membrane flavoprotein, quinone oxidoreductase (SQOR). This catalytic reaction is considered a potential drug target. To reveal the molecular basis of silane sulfur transfer reaction catalyzed by SQOR enzyme and receptors with different structures, scientists reported the crystal structure of human SQOR using an X-ray crystallography technical report. The experimental design indicates that the human SQOR contains an electropositive surface depression, a unique feature believed to be the critical point of sulfide acceptors. It is speculated that the depressed site is connected to the hydrophobic internal tunnel that binds coenzyme Q to facilitate the transport of negatively charged substrates to the hydrophilic H2S oxidation active site. These findings support a proposed catalytic model and open the door to structure-based drug design.
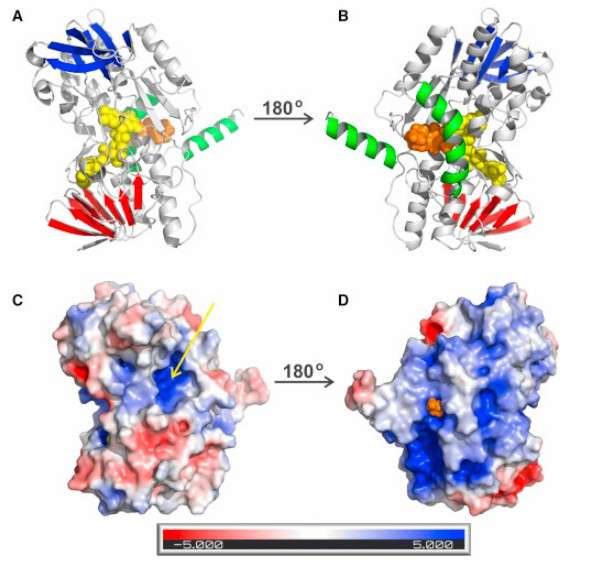 Figure 2. Overall Structure of Human SQOR. (JACKSON, et al., 2019)
Figure 2. Overall Structure of Human SQOR. (JACKSON, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Quinone oxidoreductase in complex with decylubiquinone | Aquifex aeolicus | X-ray diffraction | 2.00 Å | 3HYW |
| Quinone oxidoreductase "as-purified" protein | Aquifex aeolicus | X-ray diffraction | 2.30 Å | 3HYV |
| Quinone oxidoreductase in complex with aurachin C | Aquifex aeolicus | X-ray diffraction | 2.9 Å | 3HYX |
| Quinone oxidoreductase, selenomethionine-substituted | Homo sapiens | X-ray diffraction | 2.59 Å | 6MO6 |
| Quinone oxidoreductase, native protein | Homo sapiens | X-ray diffraction | 2.99 Å | 6MP5 |
| ETF-QO with bound UQ | Sus scrofa | X-ray diffraction | 2.5 Å | 2GMH |
| ETF-QO UQ-free structure | Sus scrofa | X-ray diffraction | 2.6 Å | 2GMJ |
| Mitochondrial trifunctional protein | Homo sapiens | X-ray diffraction | 3.6 Å | 6DV2 |
| Electron bifurcating flavoprotein Fix/EtfABCX | Thermotoga maritima MSB8 | Cryo-EM | 2.90 Å | 7KOE |
| GlpD, native | Escherichia coli | X-ray diffraction | 1.75 Å | 2QCU |
| SeMet-GlpD | Escherichia coli | X-ray diffraction | 1.95 Å | 2R4J |
| GlpD-2-PGA | Escherichia coli | X-ray diffraction | 2.3 Å | 2R45 |
| GlpD-PEP | Escherichia coli | X-ray diffraction | 2.1 Å | 2R46 |
| GlpD-DHAP | Escherichia coli | X-ray diffraction | 2.1 Å | 2R4E |
Table 1. Structural research of oxidoreductases.
Creative Biostructure specializes in membrane protein analysis services based on X-ray crystallography and cryo-electron microscopy (cryo-EM) techniques, providing high quality, efficient and reliable analytical solutions to our clients. We have a team of industry veterans who combine the most advanced analytical methods to provide our clients a full range of membrane protein analysis services. Whether you are engaged in drug discovery, or therapeutic design, or have a need for in-depth research on membrane protein structure and function, we can provide you with the highest quality services to help you achieve more prestigious achievements in related fields. Contact us to experience our professional, efficient and high-quality services.
References
- YEH J I, et al. Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proceedings of the National Academy of Sciences, 2008, 105(9): 3280–3285.
- FENG X, et al. Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein fix/etfabcx. Proceedings of the National Academy of Sciences, 2020, 118(2).
- XIA C, et al. Crystal structure of human mitochondrial trifunctional protein, a fatty acid β-oxidation metabolon. Proceedings of the National Academy of Sciences, 2019, 116(13): 6069–6074.
- ZHANG J, et al. Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool. Proceedings of the National Academy of Sciences, 2006, 103(44): 16212–16217.
- JACKSON M R, et al. X-ray structure of human sulfide:quinone oxidoreductase: Insights into the mechanism of mitochondrial hydrogen sulfide oxidation. Structure, 2019, 27(5).
- MARCIA M, et al. TThe structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proceedings of the National Academy of Sciences, 2009, 106(24): 9625–9630.
