Structural Research of Potassium, Sodium, & Proton Ion-Selective Channels
Protein dynamics are essential for organismal function, and among the widely researched are the various gating mechanisms of ion channels. Ion channels mediate the selective passive transport of ions across biological membranes, acting as transmembrane proteins that provide barriers to the passage of components. Common ion channels include potassium, sodium, and proton ion-selective channels. Unraveling the structure of ion channels is essential for solving practical problems such as drug design, channelopathies, and acquired insecticide resistance.
Advances in research on the structure of potassium ion-selective channels
Research has shown that the signature sequence for potassium ion selectivity is the TVGYG sequence, which forms a selective filter for potassium ion channels. The first potassium ion channel structure is resolved from Streptomyces lividans and named KcsA. In KcsA, the average coordination distance between oxygen and potassium ions is 2.85 Å. The structure reveals that KcsA is a tetramer consisting of four identical polypeptide chains that form a symmetric structure. In the center is a pore with conserved residues arranged to form a selective filter.
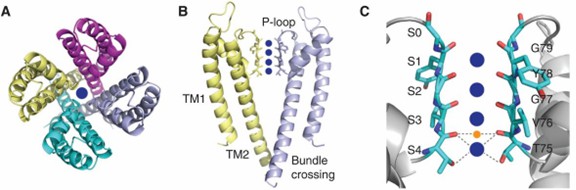 Figure 1. Structural details of KcsA. (Kim DM, Nimigean CM, 2016)
Figure 1. Structural details of KcsA. (Kim DM, Nimigean CM, 2016)
Action mechanism and structure of sodium ion-selective channels
Voltage-gated sodium channels initiate action potentials in nerves, muscles, and other excitable cells. Na+ is partially dehydrated by interacting with high-field sites containing carboxyl side chains at the outer end of the pore, and then rehydrated in the pore lumen and enters the pore. The researchers determined the crystal structure of the bacterial sodium channel NaVAb at 2.7 Å and found that NaVAb has a central pore surrounded by four pore-forming modules consisting of S5 and S6 segments as well as interstitial pore loops. This pore structure reveals the structural basis for confined ions and drug-gated entry into the pore lumen observed in studies of ion selectivity and pore closure.
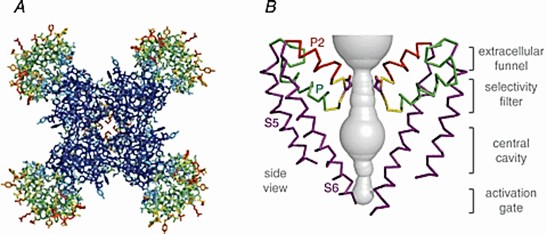 Figure 2. Structure of the NaVAb channel. (Catterall WA, 2014)
Figure 2. Structure of the NaVAb channel. (Catterall WA, 2014)
Structure-based action mechanism of potassium ion-selective channels
The voltage-gated proton channel HV1 plays a critical role in health and disease in different tissues and species. Researchers have combined biochemical, computational, and electron paramagnetic resonance (EPR) spectroscopic methods to reveal the structure of the proton channel HV1. Arg near Asp in the selectivity filter (SF) of the open HV1 channel is the narrow region that determines selectivity. The Asp and Arg side chains are attached to a ring scaffold, forming a double-toothed hydrogen bond that closes the pore. Introducing H3O+ protonates the SF, breaking the Asp-Arg bond and opening the conduction pathway.
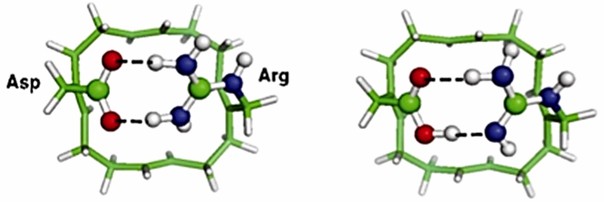 Figure 3. The ion-free Asp––Arg+ SF model. (Dudev T, et al., 2015)
Figure 3. The ion-free Asp––Arg+ SF model. (Dudev T, et al., 2015)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Potassium channel (KcsA) full-length fold | Streptomyces lividans | SOLUTION NMR | / | 1F6G |
| Potassium channel (KcsA) open gate model | Streptomyces lividans | SOLUTION NMR | / | 1JQ1 |
| KvAP | Aeropyrum pernix | Cryo-EM single particle analysis | 5.9 Å | 6UWM |
| KcsA Open gate E71V mutant in Potassium | Streptomyces lividans | X-ray diffraction | 3 Å | 7MUB |
| KcsA potassium channel with TBA and potassium | Streptomyces lividans | X-ray diffraction | 2.8 Å | 1J95 |
| Potassium channel KcsA-Fab complex in Rb+ | Mus musculus | X-ray diffraction | 2.4 Å | 1R3I |
| Mutant KcsA potassium channel | Mus musculus | X-ray diffraction | 2.5 Å | 1ZWI |
| MthK pore crystallized in the presence of TBSb | Methanothermobacter thermautotrophicus str. Delta H | X-ray diffraction | 1.7 Å | 4HZ3 |
| SthK cyclic nucleotide-gated potassium channel | Spirochaeta thermophila DSM 6578 | Cryo-EM single particle analysis | 3.42 Å | 6CJQ |
| KvAP-33H1 Fv complex | Aeropyrum pernix | X-ray diffraction | 3.9 Å | 2A0L |
| NaChBac in GDN | Halalkalibacterium halodurans C-125 | Cryo-EM single particle analysis | 3.7 Å | 6VX3 |
| NaChBac in lipid nanodisc | Halalkalibacterium halodurans C-125 | Cryo-EM single particle analysis | 3.1 Å | 6VWX |
| Sodium channel in high calcium, I222 space group, crystal 2 | Alkalilimnicola ehrlichii | X-ray diffraction | 3.8 Å | 4LTP |
| NavAb Voltage-gated Sodium Channel, residues 1-239 | Aliarcobacter butzleri RM4018 | X-ray diffraction | 2.4 Å | 6MVA |
| NavAb voltage-gated sodium channel, I217C/F203A | Aliarcobacter butzleri RM4018 | X-ray diffraction | 2.9 Å | 6MVV |
| NavAb voltage-gated sodium channel | Aliarcobacter butzleri RM4018 | X-ray diffraction | 3.21 Å | 4EKW |
| NavRh, a voltage-gated sodium channel | alpha proteobacterium HIMB114 | X-ray diffraction | 3.052 Å | 4DXW |
| NavAb voltage-gated sodium channel in an open state | Aliarcobacter butzleri | X-ray diffraction | 2.85 Å | 5VB8 |
| NavAb voltage-gated sodium channel | Aliarcobacter butzleri RM4018 | X-ray diffraction | 2.8 Å | 3RVZ |
Table 1. Structural research of potassium, sodium, and proton ion-selective channels.
Creative Biostructure is a cutting-edge biomolecule structure elucidation company offering a range of techniques for determining the 3D structure of proteins, such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy. our experienced researchers can determine the structure of ion channels and gain insights into their molecular mechanisms.
As a leader in protein structural analysis services, we are committed to advancing structural biology research and providing high-quality services to our clients. If you have requirements, please feel free to contact us for more information.
References
- Kim DM, Nimigean CM. Voltage-Gated Potassium Channels: A Structural Examination of Selectivity and Gating. Cold Spring Harb Perspect Biol. 2016.8(5): a029231.
- Catterall WA. Structure and function of voltage-gated sodium channels at atomic resolution. Exp Physiol. 2014.99(1):35-51.
- Dudev T,et al. Selectivity Mechanism of the Voltage-gated Proton Channel, HV1. Sci Rep. 2015.5:10320.
- Langan PS, et al. The structure of a potassium-selective ion channel reveals a hydrophobic gate regulating ion permeation. IUCrJ. 2020.7(Pt 5):835-843.
- Hendriks K, et al. Structural Plasticity of the Selectivity Filter in Cation Channels. Front Physiol. 2021.12:792958.