Structural Research of Insig Proteins
Cholesterol homeostasis is a finely regulated process vital for cellular function and overall health. One of the key players in this pathway is the sterol regulatory element-binding protein (SREBP) pathway, which is tightly controlled by the SREBPs and their associated proteins, Insig-1 and Insig-2. Insig proteins serve as critical regulators that monitor the sterol level in the endoplasmic reticulum (ER) membrane. Unraveling the structural basis of how Insig proteins interact with cholesterol and other components of the SREBP pathway has been a subject of intense research.
The N-terminal transcription factor domain and C-terminal regulatory domain (CTD) of SREBPs are connected by a transmembrane (TM) hairpin. Insig proteins, particularly Insig-2, interact with the C-terminal WD40 domain of the SREBP-cleavage activating protein (Scap). This interaction plays a key role in regulating the translocation of the SREBP-Scap complex to the Golgi, where it undergoes proteolytic cleavage, leading to the activation of genes involved in cholesterol synthesis and uptake.
Recent advancements in structural biology, particularly single-particle cryo-electron microscopy (cryo-EM), have provided powerful tools for investigating the structure and dynamics of membrane protein complexes like Insig. A notable study focused on deciphering the structure of the Scap-Insig complex bound to 25-hydroxycholesterol (25HC), shedding light on the molecular basis of sterol sensing.
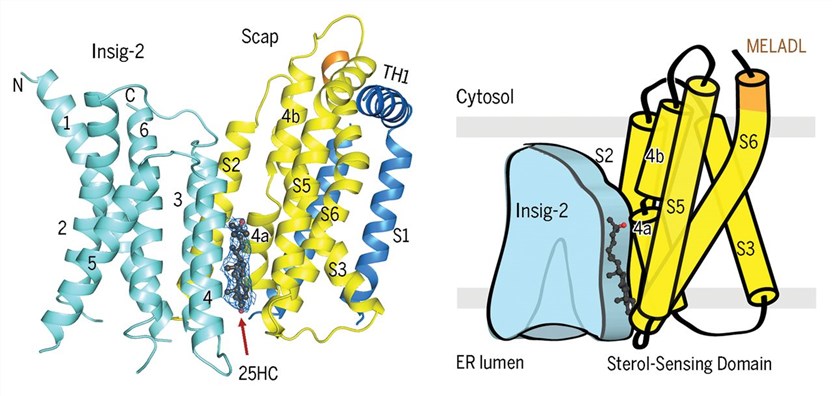 Figure 1. Cryo-EM structure of the complex of Scap and Insig-2 in the presence of 25HC. (Yan R, et al., 2021)
Figure 1. Cryo-EM structure of the complex of Scap and Insig-2 in the presence of 25HC. (Yan R, et al., 2021)
Unveiling the Molecular Basis of Sterol Sensing
The cryo-EM structure of the Scap-Insig-25HC ternary complex has revealed crucial details about how Insig proteins sense cholesterol levels in the ER membrane. TMs 1, 2, 3, and 6 of Insig-2 form a hydrophobic pocket without any density corresponding to a sterol, whereas 25HC is perfectly accommodated between Scap and Insig-2 in the luminal leaflet of the membrane.
The binding site consists mainly of hydrophobic residues on TMs 3 and 4 of Insig-2 and S4, S5, and S6 of Scap. Notably, the 25-OH group at the end of the iso-octanol tail of 25HC is exposed to the cytosolic milieu through a hydrophilic cavity enclosed by Scap and Insig-2. This unique structural arrangement might explain the preferential interaction between 25HC and Insig, making it more sensitive to cholesterol derivatives like 25HC compared to cholesterol itself.
Crucial Role of Scap-S4 Unwinding in Sterol Sensing
The broken S4 in Scap forms two half helices, S4a and S4b, and the tilting of S4a towards the interior of the sterol-sensing domain creates space for 25HC accommodation and displacement of S2, a major interface with Insig. This unwound conformation of Scap-S4 is essential for stable Insig association, and the presence of 25HC further stabilizes this conformation. Notably, specific Scap mutations, such as D428A and Q432A, affect the unwinding of S4, allowing for complex formation with Insig in the absence of sterols.
| Protein | Organism | Method | Resolution | PDB Entry ID |
| MvINS Insig homolog (expressed in Escherichia coli) | Mycolicibacterium vanbaalenii PYR-1 | X-ray diffraction | 1.90 Å | 4XU4 |
| Scap/Insig complex in the presence of 25-hydroxyl cholesterol (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.70 Å | 6M49 |
Table 1. Structural research of Insig proteins.
Creative Biostructure stands out as a reliable partner for cutting-edge structural biology research, with a commitment to providing high-quality structural analysis services and delivering valuable insights into challenging biological questions. Our team of experts excels in utilizing advanced techniques like cryo-EM to unlock the mysteries of membrane protein complexes.
With a long-standing reputation and a track record of successful collaborations with esteemed researchers and organizations, we continue to be a leading choice for clients seeking detailed structural analysis and research support. By employing advanced methodologies, including X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM), our team of proficient scientists excels in unveiling the intricate three-dimensional architectures of membrane protein complexes. This expertise enables us to gain insights into their functional mechanisms and interactions, contributing significantly to the comprehension of biological processes. Contact us now to explore the full potential of our capabilities, which can elevate your research endeavors and propel you towards the attainment of your scientific aspirations.
References
- Ren R, et al. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels. Science. 2015, 349(6244): 187-191.
- Yan R, et al. A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols. Science. 2021, 371(6533): eabb2224.