Ribosome Structure
Ribosomes are essential molecular machines found in all living cells and are responsible for synthesizing proteins by translating messenger RNA (mRNA) into polypeptide chains. This process, known as translation, is fundamental to cellular function and growth. The structure of ribosomes is complex and highly conserved across different domains of life, including prokaryotes, archaea, eukaryotes, and organelles such as mitochondria and chloroplasts. Understanding the structure of ribosomes is important for several reasons, ranging from basic biological research to practical applications in medicine and biotechnology. This knowledge impacts our understanding of fundamental life processes, informs drug design, and enhances our ability to manipulate biological systems for various applications.
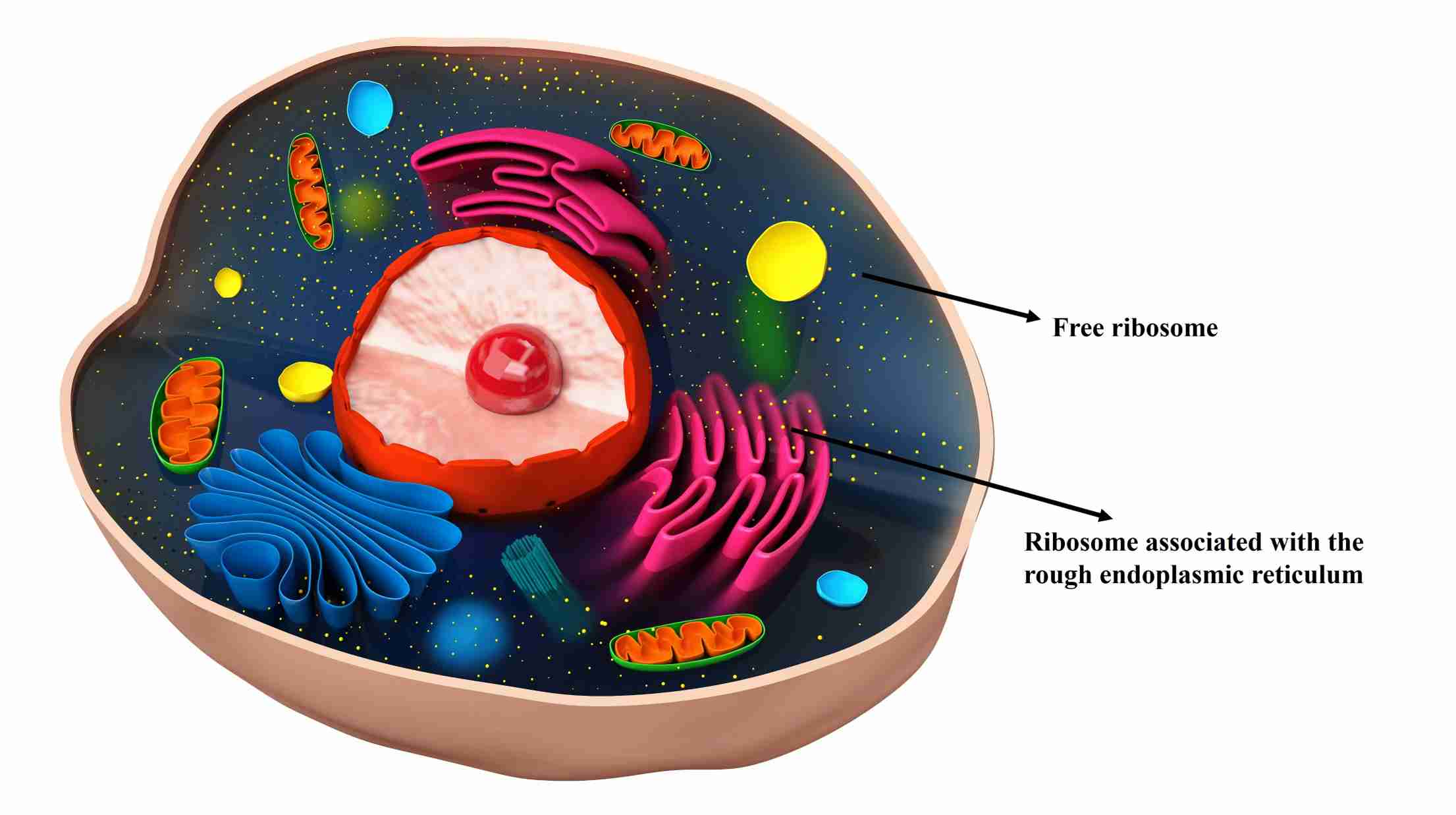 Figure 1. Illustration of free ribosome and bound ribosome in eukaryotic cell.
Figure 1. Illustration of free ribosome and bound ribosome in eukaryotic cell.
Ribosomal Subunits
Ribosomes are granular particles about 20-25 nm in diameter. They consist of about two-thirds ribosomal RNA (rRNA) and one-third ribosomal proteins. In all organisms, they are composed of two differently sized and functionally distinct subunits. The mass of ribosomes is characterized by their sedimentation behavior, which is expressed in Svedberg units (S). During translation, they assemble into a functional complex in which the large subunit (LSU) links the amino acids into a chain in protein biosynthesis (peptidyl transferase activity) and the small subunit (SSU) is responsible for mRNA recognition. Both subunits consist of proteins and rRNA, with the proteins responsible for cohesion and correct positioning, while the actual reactions are carried out by the rRNAs. In eukaryotes, both subunits are formed in the nucleoli of the cell nucleus and are then released into the cytoplasm through the nuclear pores.
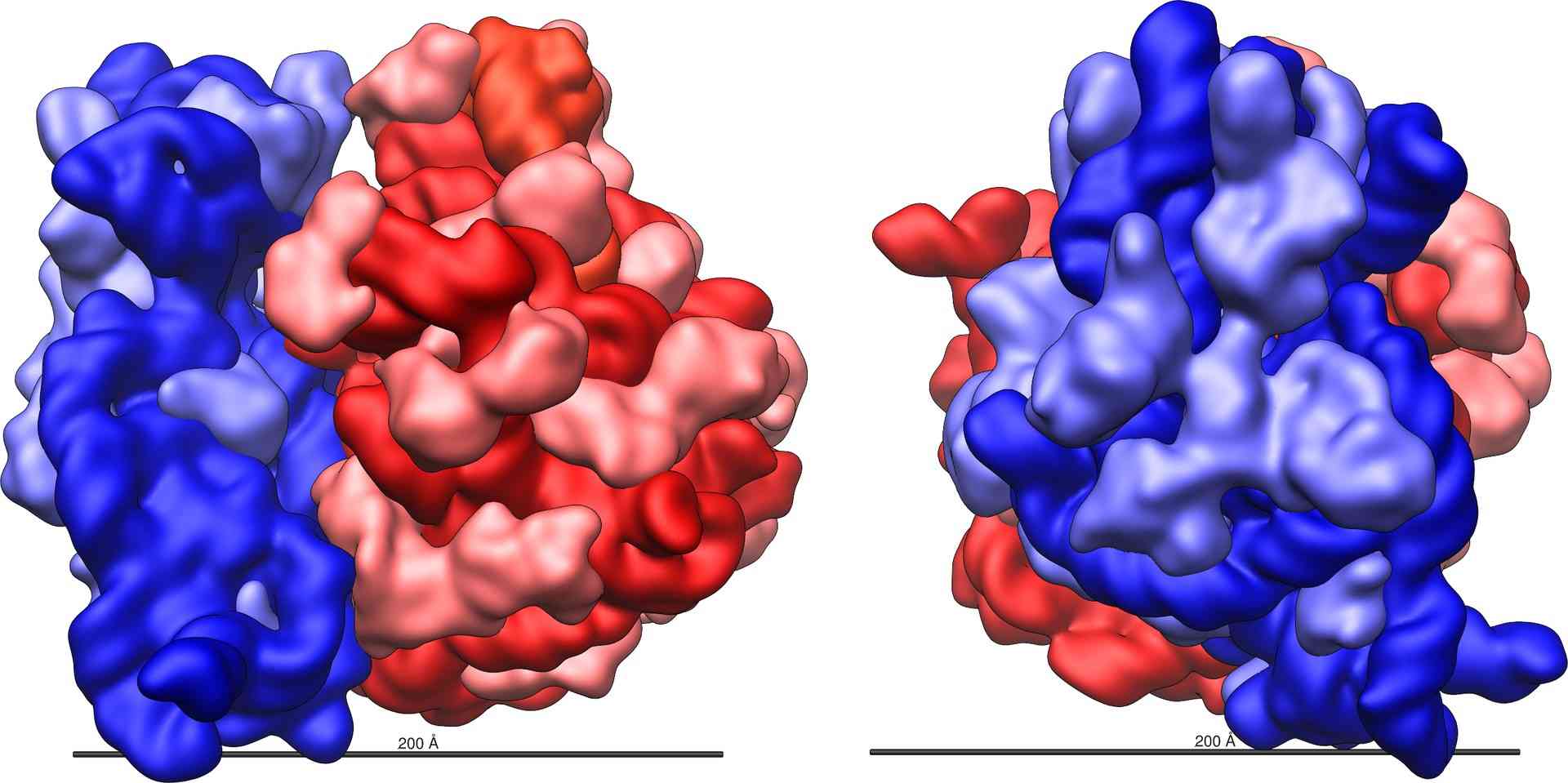 Figure 2. Large (red) and small (blue) subunits of a ribosome.
Figure 2. Large (red) and small (blue) subunits of a ribosome.
Types of Ribosomes
Prokaryotic Ribosomes: Prokaryotic ribosomes found in bacteria are 70S ribosomes consisting of a 30S SSU and a 50S LSU. The 30S subunit contains 16S rRNA, which plays a critical role in mRNA binding and decoding. The 50S subunit contains 23S and 5S rRNAs, the 23S rRNA being part of the peptidyl transferase center. Prokaryotic ribosomes are targets for many antibiotics that exploit differences between prokaryotic and eukaryotic ribosomes to inhibit bacterial protein synthesis without affecting human cells.
Archaeal Ribosomes: Archaeal ribosomes are also 70S ribosomes, similar in size to prokaryotic ribosomes. However, their protein composition and rRNA sequences have distinct features that are more closely related to eukaryotic ribosomes, reflecting the unique evolutionary position of archaea. Archaeal ribosomes have been studied for their stability and ability to function in extreme environments, providing insights into the adaptation of ribosomal structures to different environmental conditions.
Eukaryotic Ribosomes: Eukaryotic ribosomes are 80S ribosomes consisting of a 40S SSU and a 60S LSU. The 40S subunit contains 18S rRNA, which is essential for mRNA binding and decoding, while the 60S subunit contains 28S, 5.8S, and 5S rRNAs, with the 28S rRNA being part of the peptidyl transferase center. Eukaryotic ribosomes are more complex than prokaryotic ribosomes, reflecting the greater complexity of eukaryotic cells and their need for more sophisticated regulatory mechanisms in protein synthesis.
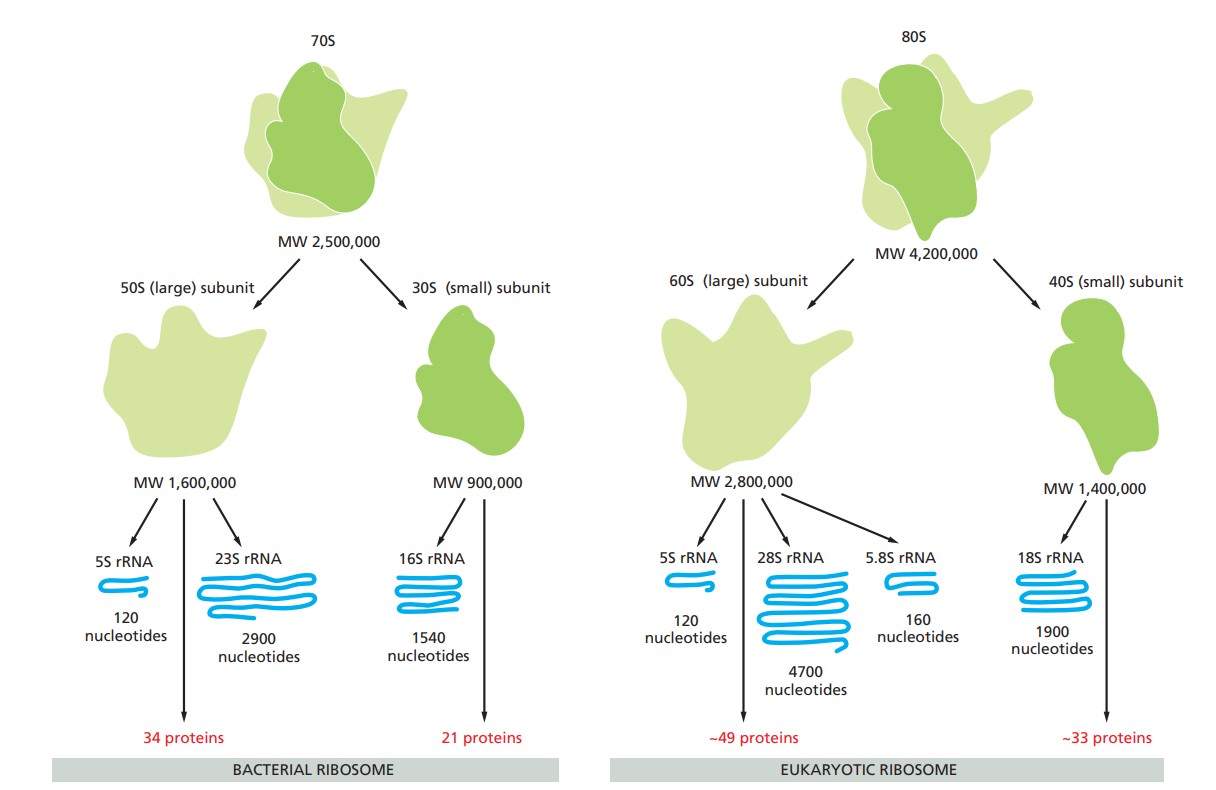 Figure 3. A comparison of bacterial and eukaryotic ribosomes (Molecular Biology of the Cell, 6th Edition).
Figure 3. A comparison of bacterial and eukaryotic ribosomes (Molecular Biology of the Cell, 6th Edition).
Plastoribosomes and Mitoribosomes
Plastoribosomes are found in chloroplasts and are similar to prokaryotic ribosomes, reflecting the endosymbiotic origin of chloroplasts from cyanobacteria. Plastoribosomes are involved in the synthesis of proteins essential for photosynthesis and other chloroplast functions.
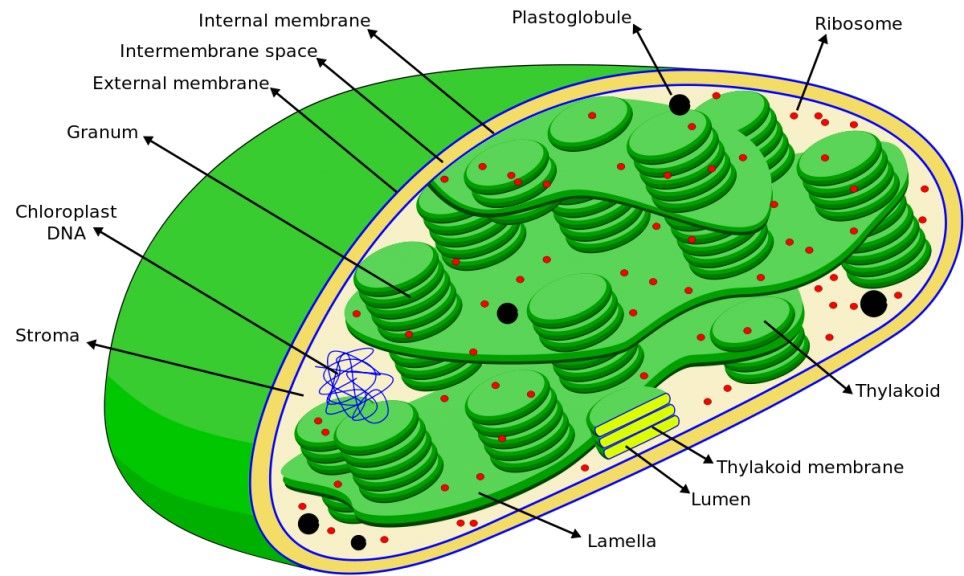 Figure 4. A diagram showing plastoribosomes among other chloroplast structures.
Figure 4. A diagram showing plastoribosomes among other chloroplast structures.
Mitoribosomes, specialized ribosomes found in mitochondria, play a critical role in the translation of mitochondrial DNA-encoded proteins essential for the function of the organelle. These ribosomes exhibit remarkable structural diversity across species, reflecting the unique evolutionary pressures and functional requirements of each organism.
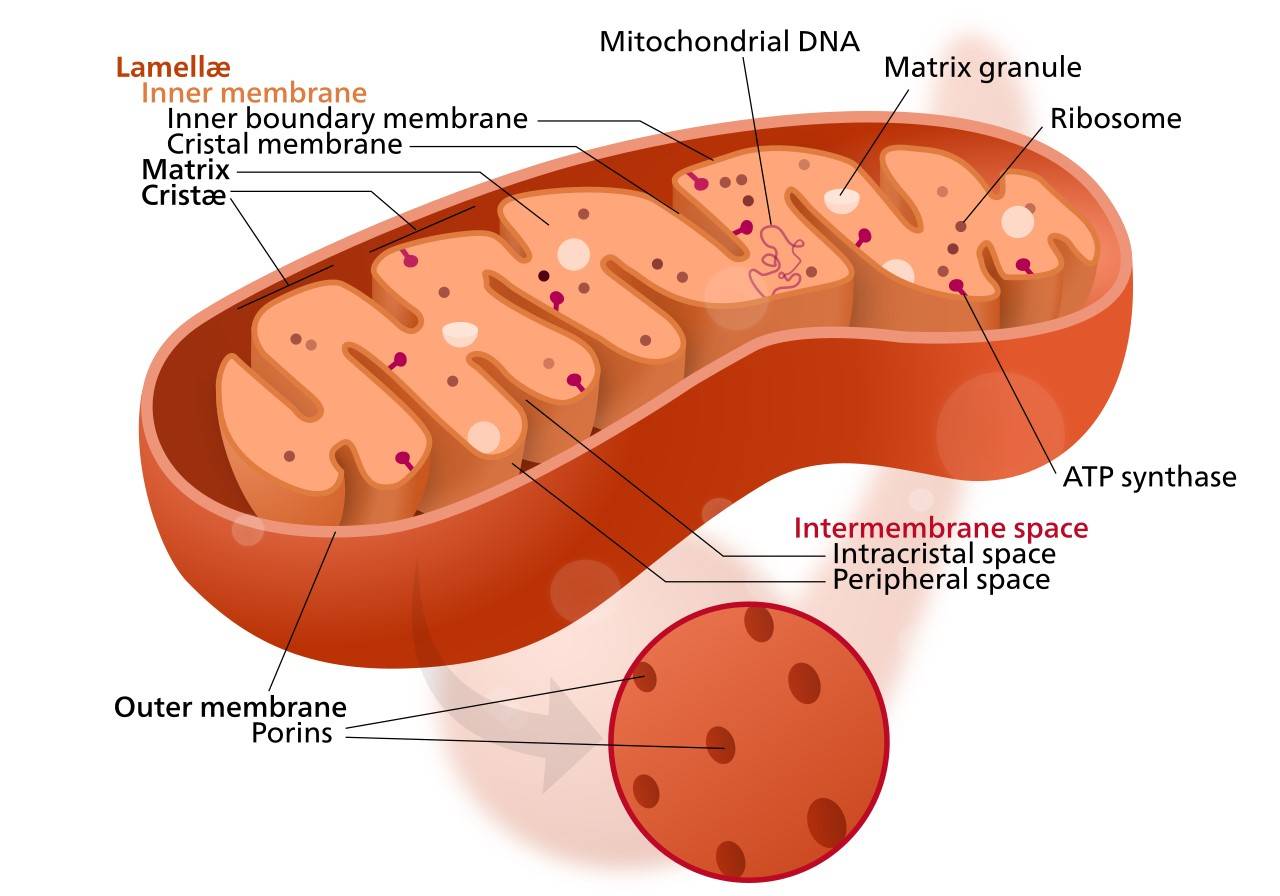 Figure 5. A diagram showing mitoribosomes among other mitochondria structures.
Figure 5. A diagram showing mitoribosomes among other mitochondria structures.
Unlike cytoplasmic ribosomes, mitoribosomes contain specialized ribosomal RNA (rRNA) and protein components. These components are highly adapted to the specialized needs of the mitochondrion, particularly for oxidative phosphorylation, the process by which ATP is produced. The unique rRNA sequences and incorporation of mitochondria-specific proteins enable mitoribosomes to efficiently synthesize the 13 core proteins of the electron transport chain, as well as other key proteins involved in mitochondrial metabolic pathways. This structural and functional specialization underscores the critical role of the mitoribosome in cellular energy production and highlights its evolutionary adaptation to the unique environment of the mitochondrion.
In human, for example, mitoribosomes are classified as 55S, composed of a 28S small subunit (SSU) and a 39S large subunit (LSU). In detail, it consists of three rRNA molecules (16S mt-LSU rRNA, 12S mt-SSU rRNA, and mt-tRNAVal) and 80 proteins, 36 of which are specific to mitochondria. In addition, most proteins with homologs in bacteria have substantial extensions. The increased protein mass results in a ribosome with a unique morphology, a more complex network of over 200 protein-protein interactions, and an rRNA core that is more effectively protected from reactive oxygen species.
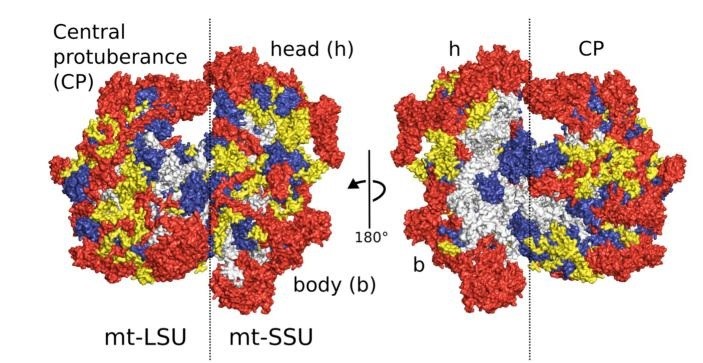 Figure 6. Overview of the human mitoribosome. Proteins conserved with bacteria (blue), extensions of homologous proteins (yellow) and mitochondria-specific proteins (red). rRNA is shown in gray (Amunts et al., 2015).
Figure 6. Overview of the human mitoribosome. Proteins conserved with bacteria (blue), extensions of homologous proteins (yellow) and mitochondria-specific proteins (red). rRNA is shown in gray (Amunts et al., 2015).
Methods for Studying Ribosome Structure
A thorough comprehension of ribosomal structure is indispensable for an accurate understanding of its function and mechanisms. A variety of methods are utilized for the analysis of ribosomal structure, including:
| Techniques | Description |
| X-ray Crystallography | X-ray crystallography reveals high-resolution ribosome structures by analyzing diffraction patterns of crystallized ribosomal subunits, elucidating active sites, tRNA binding, and antibiotic interactions. |
| Cryo-Electron Microscopy (Cryo-EM) | Cryo-EM enables near-atomic visualization of ribosomes without crystallization by freezing samples and using advanced image processing to reconstruct detailed 3D structures and functional states. |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | NMR spectroscopy reveals ribosomal dynamics and conformational changes, especially useful for studying smaller subunits, rRNA, and proteins, complementing data from other structural techniques. |
| Mass Spectrometry (MS) | Mass spectrometry is a highly sensitive and quantitative method for determining the binding kinetics of r-proteins to rRNA and for studying when multiple r-protein or rRNA chemical modifications are introduced during assembly. |
| Pulse-Chase Quantitative Mass Spectrometry (PC-qMS) | PC-qMS allows tracking the binding rates of all r-proteins to rRNA in a single experiment. qMS-based methods have also been used to recapitulate the assembly pathway in vivo and to identify multiple assembly factors such as GTPases, helicases, and maturation factors. |
| Cross-linking and Mass Spectrometry | CL-MS studies ribosome interactions by linking and identifying interacting protein and RNA regions, mapping ribosomal structure and functional domains through spatial proximity and interaction networks. |
| Molecular Dynamics Simulations | MD simulations provide insight into the dynamic behavior of ribosomes at the atomic level. These simulations model the motion and interactions of ribosomal components over time, helping to understand conformational changes, binding events, and the effects of mutations. MD simulations complement experimental data to provide a deeper understanding of ribosomal function and regulation. |
| Fluorescent Resonance Energy Transfer (FRET) | FRET provides insights into the spatial relationships and interactions within ribosomes and can be used for studying following translocation and ribosome dynamics. |
| Single Molecular Polarized Total Internal Reflection Fluorescence (polTIRF) Microscopy | polTIRF provides information not only on the distance but also on the relative orientation between domains. By applying this method to the ribosome, the researchers were able to observe a power stroke of domain IV of EF-G during translocation. |
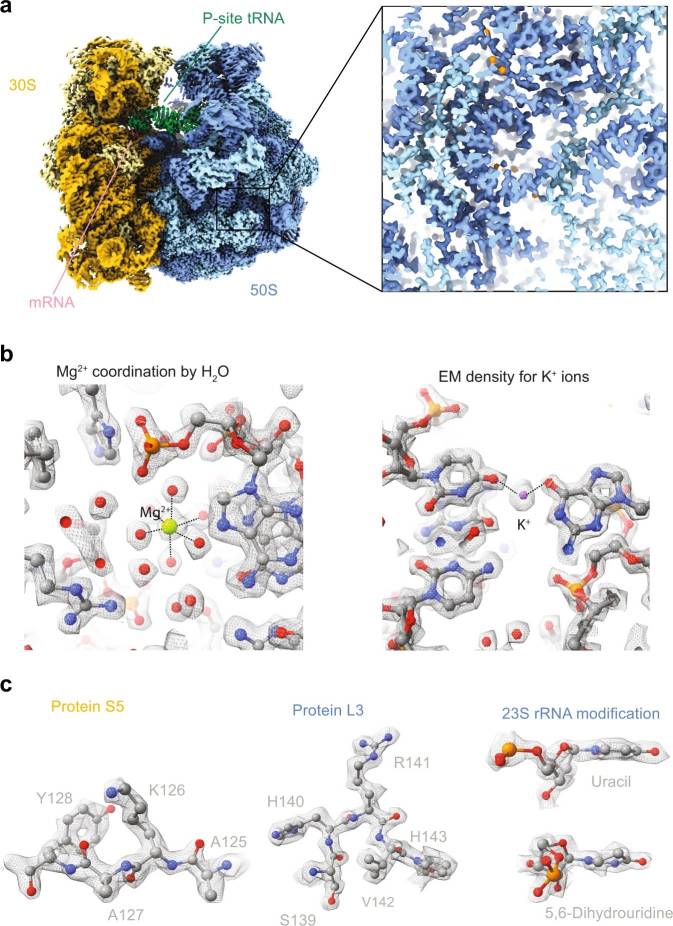 Figure 7. Example of a cryo-EM study of a translating ribosome from Escherichia coli (Fromm et al., 2023).
Figure 7. Example of a cryo-EM study of a translating ribosome from Escherichia coli (Fromm et al., 2023).
Ribosomes are fundamental to cellular life, synthesizing proteins based on genetic instructions encoded in mRNA. The intricate structure of ribosomes, composed of rRNA and proteins, enables them to perform this vital function with high precision and efficiency. Despite their ubiquity, ribosomes exhibit structural variation in different domains of life and within organelles, reflecting evolutionary adaptations to different cellular environments.
Creative Biostructure is dedicated to providing advanced cryo-EM solutions for ribosome structures, which play a critical role in cellular biology and drug discovery and development. For more information on how our services can support your research, please contact us for a customized quote.
References
- Molecular Biology of the Cell, 6th edition.
- Amunts, A., Brown, A., Toots, J., Scheres, S. H. W., & Ramakrishnan, V. (2015). The structure of the human mitochondrial ribosome. Science (New York, N.Y.), 348(6230), 95–98.
- Fromm, S. A., O'Connor, K. M., Purdy, M., Bhatt, P. R., Loughran, G., Atkins, J. F., Jomaa, A., & Mattei, S. (2023). The translating bacterial ribosome at 1.55 Å resolution generated by cryo-EM imaging services. Nature Communications, 14(1), 1095.
- Gor, K., & Duss, O. (2023). Emerging quantitative biochemical, structural, and biophysical methods for studying ribosome and protein–rna complex assembly. Biomolecules, 13(5), 866.
- Jobe, A., Liu, Z., Gutierrez-Vargas, C., & Frank, J. (2019). New insights into ribosome structure and function. Cold Spring Harbor Perspectives in Biology, 11(1), a032615.
- Lai, C. (Wan-J., & Ermolenko, D. N. (2018). Ensemble and single-molecule FRET studies of protein synthesis. Methods (San Diego, Calif.), 137, 37–48.
