Structural Research of Antiporters
Antiporters are a type of membrane transport protein that can move two different types of molecules or ions in opposite directions across a cell membrane. Structural research on antiporters has progressed rapidly in recent years, aided by the development of advanced techniques such as X-ray crystallography, cryo-electron microscopy, and molecular dynamics simulations.
Some of the key findings from structural research on antiporters include the identification of specific amino acid residues that are critical for their function, the elucidation of their transport mechanism, and the discovery of novel antiporter structures with distinct properties.
For example, recent studies have revealed the crystal structures of several antiporters, including the lactose permease, the sodium/proton exchanger NhaA, and the chloride/proton exchanger ClC-ec1. These structures have provided valuable insights into the molecular basis of antiporter function, such as how they recognize and bind their substrates, and how they undergo conformational changes to transport them across the membrane.
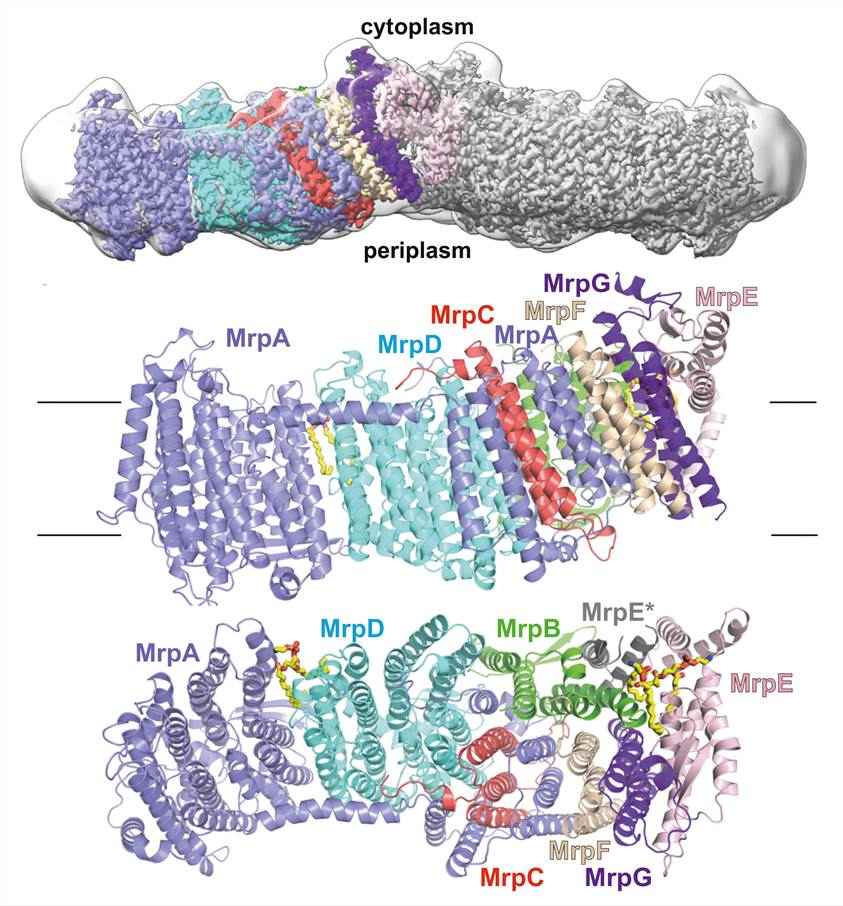 Figure 1. Overall structure of Mrp Na+/H+ antiporter. (Lee Y, et al., 2022)
Figure 1. Overall structure of Mrp Na+/H+ antiporter. (Lee Y, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| NhaA Na+/H+ antiporter (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.45 Å | 1ZCD |
| NhaA dimer model | Escherichia coli | Electron Crystallography | 7.00 Å | 3FI1 |
| NhaA dimer, crystallised at low pH (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.70 Å | 4AU5 |
| Na+/H+ antiporter NhaA at pH 6.5 (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.20 Å | 7S24 |
| NapA Na+/H+ antiporter (expressed in E. coli) | Thermus thermophilus | X-ray diffraction | 2.98 Å | 4BWZ |
| NapA Na+/H+ antiporter, outward facing (expressed in E. coli) | Thermus thermophilus | X-ray diffraction | 2.30 Å | 5BZ3 |
| NhaP Na+/H+ antiporter, pH 8 (expressed in E. coli) | Pyrococcus abyssi | X-ray diffraction | 3.15 Å | 4CZ8 |
| NhaP1 Na+/H+ antiporter, pH 8 (expressed in E. coli) | Methanocaldococcus jannaschii | X-ray diffraction | 3.50 Å | 4CZB |
| Mitochondrial ADP/ATP carrier in complex with carboxyatractyloside | Bos taurus | X-ray diffraction | 2.20 Å | 1OKC |
| Mitochondrial ADP/ATP Carrier | Bos taurus | X-ray diffraction | 2.80 Å | 2C3E |
| Mitochondrial ADP/ATP carrier isoform 2 inhibited by carboxyatractyloside (C2221 crystal form) (expressed in Saccharomyces cerevisiae) | Saccharomyces cerevisiae | X-ray diffraction | 2.49 Å | 4C9G |
| Mitochondrial ADP/ATP carrier, bongkrekic acid-inhibited (expressed in Saccharomyces cerevisiae) | Thermothelomyces thermophilus | X-ray diffraction | 3.30 Å | 6GCI |
| UCP2 mitochondrial uncoupling protein 2 (expressed in E. coli) | Mus musculus | Solution NMR | / | 2LCK |
| Multiple resistance & pH adaptation (Mrp) complex Na+/H+ antiporter (expressed in E. coli) | Dietzia sp. DQ12-45-1b | Cryo-EM single particle analysis | 3.00 Å | 7D3U |
| Multiple resistance & pH adaptation (Mrp) complex Na+/H+ antiporter (expressed in E. coli) | Alkalihalobacillus pseudofirmus | Cryo-EM single particle analysis | 2.24 Å | 7QRU |
| Na+/H+ exchanger NHE9 (SLC9A9), inward-facing conformation (expressed in Saccharomyces cerevisiae) | Equus caballus | Cryo-EM single particle analysis | 3.51 Å | 6Z3Y |
| Na+/H+ exchanger NHA2 (SLC9B2) in detergent with N-terminal extension helix (expressed in Saccharomyces cerevisiae) | Bison bison | Cryo-EM single particle analysis | 3.15 Å | 7P1I |
| Na+/H+ exchanger NHE1-CHP1 complex, under pH 7.5 (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.30 Å | 7DSW |
Table 1. Structural Research of Antiporters.
At Creative Biostructure, we specialize in providing high-quality structural biology solutions to support drug discovery, protein engineering, and basic research. Our team of experienced scientists is skilled in a wide range of techniques, such as X-ray crystallography, electron crystallography, cryo-EM single-particle analysis and protein NMR spectroscopy.
Our services are tailored to meet your specific needs, whether you require full-service structural biology support, project-specific expertise, or technology transfer to your own team. We are committed to delivering timely, accurate results and maintaining open communication throughout the project. Please do not hesitate to contact us with any questions or to discuss your project in more detail.
References
- Winkelmann I, et al. Crystal structure of the Na+/H+ antiporter NhaA at active pH reveals the mechanistic basis for pH sensing. Nature Communications. 2022, 13(1): 6383.
- Lee Y, et al. Ion transfer mechanisms in Mrp-type antiporters from high resolution cryoEM and molecular dynamics simulations. Nature Communications. 2022, 13(1): 6091.
- Matsuoka R, et al. Structure, mechanism and lipid-mediated remodeling of the mammalian Na+/H+ exchanger NHA2. Nature Structural & Molecular Biology. 2022, 29(2): 108-120.