Structural Research of Bluetongue Virus
Bluetongue virus (BTV) is an insect-mediated pathogen of wild ruminants and livestock in many parts of the world, often causing significant economic losses to the livestock industry. BTV is a double-stranded RNA (dsRNA) virus belonging to the genus Orbivirus in the family Reoviridae, which is characterized by endogenous RNA transcription via an RNA-dependent RNA polymerase (RdRp). In recent years, a new study has revealed the atomic structure of BTV, including the way it infects healthy host cells. Scientists hope to use this information to help develop vaccines and drug treatments for bluetongue.
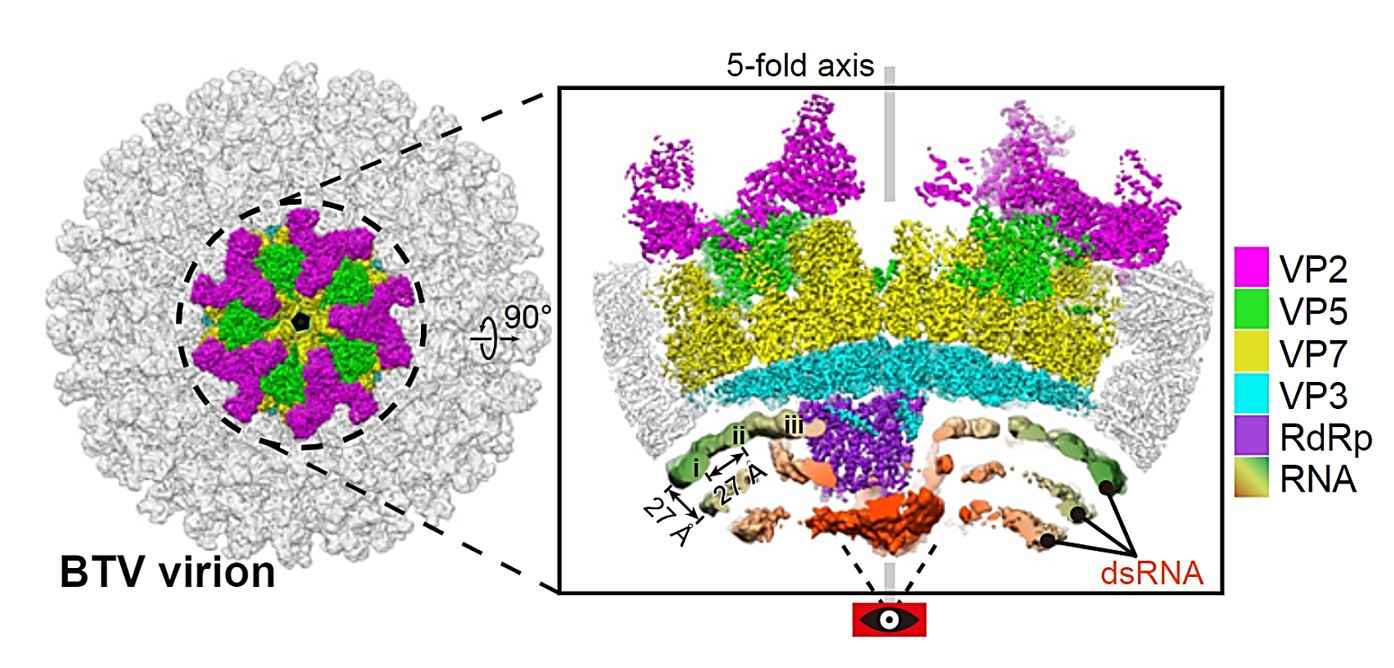 Figure 1. CryoEM reconstruction of the vertex region of BTV virion. (He Y, et al., 2019)
Figure 1. CryoEM reconstruction of the vertex region of BTV virion. (He Y, et al., 2019)
Structural Features of BTV
BTV is an enveloped virus whose genome contains 9 to 12 linear double-stranded RNA (dsRNA) fragments enclosed in an icosahedral capsid with multiple copies of RdRp. The icosahedral structure of the BTV virion and core has been determined to be of near-atomic resolution by X-ray crystallography and cryo-electron microscopy (cryo-EM). The structure shows that BTV has a three-layered capsid: an outer layer consisting of 60 stranded VP2 trimers and 120 interspersed VP5 globular trimers, an intermediate layer consisting of 260 VP7 trimers, and an inner layer formed by 60 dimers of the capsid protein VP3. Ten genomic dsRNA fragments with enzymatic proteins are within the capsid, including putative deconjugase VP6, mRNA capping enzyme VP4, and RdRp VP1.
Advances in Structural Research on BTV
BTV has been researched as a model for large envelope-free dsRNA viruses. Several new techniques have been applied to define the virally encoded enzymes required for RNA replication, thus providing the sequence for capsid assembly and its required proteins. In addition, reconstructed in vitro systems define the individual steps of genomic RNA assembly. These findings point to possible routes of infestation of related viruses and provide a basis for disease prevention. Since RNA-based genomic transcription of BTV during infection depends on RdRp, in recent years, scientists have obtained BTV RdRp structures at 3.7 Å resolution by cryo-EM and reconstruction methods, revealing how RdRp interacts with the associated genomic dsRNAs and capsid proteins.
Creative Biostructure is dedicated to providing a comprehensive range of virus-like particles (VLPs) products and viral structure analysis solutions for viral structural biology research. Our VLPs are self-assembled from viral capsid proteins that closely resemble real viruses, are easily recognized by the immune system, and present viral antigens in a pathway similar to their true conformation to induce a strong immune response, but do not contain viral genetic material, making them a safe, non-infectious tool for vaccine and antiviral drug development.
| Cat No. | Product Name | Virus Family | Source | Composition |
| CBS-V555 | Bluetongue virus VLP (VP2; VP5; VP7; VP3 Proteins) | Reoviridae | Insect cell recombinant | VP2; VP5; VP7; VP3 |
| CBS-V556 | Bluetongue virus serotype 8 VLP (VP2; VP5; VP7; VP3 Proteins) | Reoviridae | Plant recombinant | VP2; VP5; VP7; VP3 |
| Explore All Bluetongue Virus VLP Products | ||||
Creative Biostructure is a scientific research and services company specializing in viral structural biology. We offer various technologies and services including X-ray crystallography and cryo-EM. Our team of professionals has the experience and expertise to provide a full range of support and solutions for your research.
We will provide high-quality data and analytical results based on the client's requirements and research objectives, helping to gain insight into the structure of viruses and mechanisms of infestation and advancing research progress. If you are interested in our services, please contact us for a more detailed service description.
References
- He Y, et al. In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating. Proc Natl Acad Sci U S A. 2019. 116(33): 16535-16540.
- Roy P. Bluetongue virus structure and assembly. Curr Opin Virol. 2017. 24: 115-123.