Protein-Ligand Interaction
Proteins are dynamic molecules whose function is contingent upon interactions with other molecules. These interactions are influenced in physiologically significant ways by alterations in protein conformation, which can be either subtle or pronounced. The function of numerous proteins entails the reversible binding of other molecules. Molecules that bind reversibly to proteins are designated as ligands. The ligand binds to a region of the protein designated as the binding site, which is complementary in terms of size, shape, charge, and hydrophobic or hydrophilic properties.
Protein-ligand interactions are fundamental to numerous biological processes, including enzyme catalysis, signal transduction, and molecular recognition. These interactions are central to drug design and development because the binding of a ligand to a protein can modulate its function. Understanding the nature and mechanics of these interactions allows scientists to design more effective drugs, understand disease mechanisms, and explore novel therapeutic strategies.
Creative Biostructure provides high-quality assay services and customized solution including X-ray crystallography, NMR spectroscopy, surface plasmon resonance (SPR), isothermal titration calorimetry (ITC) etc.
Characteristics of Protein-Ligand Interactions
Specificity
Specificity in protein-ligand interactions refers to the ability of a protein to bind selectively to a particular ligand among a large number of potential molecules. This selectivity ensures that the protein performs its intended biological function correctly by interacting only with the correct ligand. Key factors that contribute to specificity include molecular complementarity, binding site architecture, binding site architecture, etc.
Affinity
The function of individual proteins and the interactions they belong to can be described by three factors: the amount of protein and ligand present, the extent of complex formation, and the rate of complex formation and disassembly. In conclusion, the interaction between a protein (P) and a ligand (L) to form a protein-ligand complex (PL) can be described by the equilibrium equation:
P+L ↔ PL
This equilibrium is characterized by the association constant (Ka) and the dissociation constant (Kd). The association constant Ka is defined as:
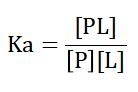
where [PL], [P], and [L] represent the concentrations of the protein-ligand complex, free protein, and free ligand, respectively. A higher Ka indicates a higher affinity between the protein and the ligand. Conversely, the dissociation constant Kd, which is the inverse of Ka, represents the ligand concentration at which half of the protein binding sites are occupied. A lower Kd indicates higher affinity, meaning that the protein and ligand are more likely to form a stable complex at lower concentrations.
Non-Covalent
In general, protein-ligand interactions are non-covalent. Non-covalent binding may depend on hydrogen bonding, hydrophobic forces, van der Waals forces, π-π interactions, electrostatic interactions in which no electrons are shared between the two or more molecules involved.
Reversible
Protein-ligand interactions are reversible binding, i.e., non-permanent interactions between a protein and a ligand in which the ligand can bind to and dissociate from the protein without any chemical change to either the protein or the ligand. This type of binding is typically governed by non-covalent interactions. Reversible binding is critical for many physiological processes, allowing dynamic regulation and control of protein function. Myoglobin and hemoglobin are perhaps the most studied and best understood proteins of reversible binding, enabling the transport and release of oxygen to tissues on demand.
The Binding Modes of Protein-Ligand Interactions
There are currently three models of protein-ligand binding - the "lock and key" model, the "induced fit" model and the "conformational selection" model.
Lock-and-Key
The lock-and-key model, first proposed by Emil Fischer in 1894, represents the simplest model of protein-ligand interaction. According to this model, the binding site of a protein is already in a perfect conformation to accept a ligand. The ligand fits into the binding site without requiring significant structural changes to the protein. The model emphasizes the pre-existing complementarity between the binding site and the ligand.
Induced Fit
The induced fit model, first proposed by Daniel Koshland in 1958, posits that the binding site of a protein does not initially align perfectly with the ligand. Instead, as the ligand approaches the protein, the protein undergoes a conformational change to achieve optimal alignment, thereby enhancing the binding interaction. The model accounts for the flexibility inherent to protein structure and recognizes that proteins are dynamic molecules.
Conformational Selection
The conformational selection model posits that proteins exist in a dynamic equilibrium between multiple conformations even in the absence of a ligand. According to this model, the ligand selects and binds the protein conformation that best fits the ligand. Binding stabilizes this particular conformation, shifting the equilibrium to the ligand-bound state. The model incorporates the idea of pre-existing conformational flexibility in proteins.
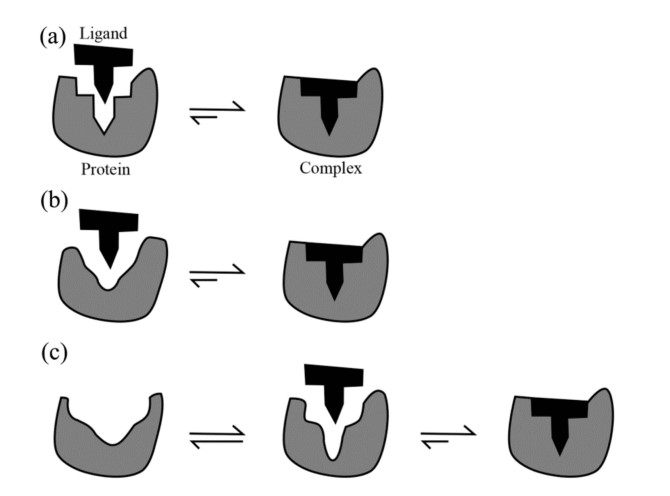 Figure 1. Schematic illustrations of the three protein-ligand binding models: (a) Lock-and-key; (b) Induced fit; and (c) Conformational selection (Du et al., 2015).
Figure 1. Schematic illustrations of the three protein-ligand binding models: (a) Lock-and-key; (b) Induced fit; and (c) Conformational selection (Du et al., 2015).
In reality, protein-ligand interactions often involve a combination of these mechanisms. A ligand may initially bind to a pre-existing conformation of the protein (conformational selection), and this interaction may further induce additional conformational changes to optimize binding (induced fit). Understanding these models provides a comprehensive framework for studying the dynamic nature of protein-ligand interactions, contributing to more effective drug design and deeper insight into biological processes.
Orthosteric vs. Allosteric: The Battle of Protein-Ligand Binding
Orthosteric Binding
Orthosteric binding refers to the interaction between a ligand and the primary active site of a protein, typically where the endogenous (naturally occurring) ligand binds. This site is often referred to as the orthosteric site.
Allosteric Binding
Allosteric binding occurs when a ligand binds to a site on the protein that is different from the active site of the protein, known as the allosteric site. This binding induces a conformational change in the protein that can either enhance or inhibit the binding of a ligand to the active site or affect the overall activity of the protein. Allosteric regulation is an important mechanism for fine-tuning protein function and is widely observed in enzymes and receptors.
Methods for Analyzing Protein-Ligand Interactions
Understanding and characterizing protein-ligand interactions requires a variety of experimental and computational methods. These methods can provide insight into the binding affinity, kinetics, and structural details of the interaction. Experimental methods are summarized in the table below, including X-ray crystallography, NMR spectroscopy, surface plasmon resonance (SPR), isothermal titration calorimetry (ITC), etc.
Computational methods for the analysis and precision of protein-ligand interactions include molecular docking, which predicts the preferred binding orientation of a ligand to a protein, and molecular dynamics (MD) simulations, which provide insight into the dynamic behavior of protein-ligand complexes over time.
| Experimental Techniques | Attribute measured |
| X-ray Crystallography | X-ray crystallography provides high-resolution, three-dimensional structures of protein-ligand complexes. |
| Nuclear Magnetic Resonance (NMR) Spectroscopy | NMR spectroscopy offers detailed information about the dynamics and interactions within protein-ligand complexes in solution. |
| Surface Plasmon Resonance (SPR) | SPR measures the binding kinetics and affinity of protein-ligand interactions in real time. |
| Isothermal Titration Calorimetry (ITC) | ITC directly measures the heat change associated with the binding process. |
Protein-Ligand Interactions in Biological Systems and Drug Discovery
Several notable examples illustrate the importance of protein-ligand interactions in biological systems and drug discovery.
Enzyme-Substrate Interactions: The binding of substrates to enzymes is essential for catalysis. For example, the interaction between the enzyme acetylcholinesterase and its substrate acetylcholine is critical for nerve signaling. Acetylcholinesterase inhibitors, such as donepezil, are used to treat Alzheimer's disease by preventing the breakdown of acetylcholine, thereby enhancing neurotransmission.
Kinase Inhibitors: Protein kinases play a critical role in cell signaling and cancer progression. Inhibitors that target specific kinases, such as imatinib for BCR-ABL kinase in chronic myeloid leukemia, exemplify the therapeutic potential of modulating protein-ligand interactions. Imatinib binds to the ATP-binding site of BCR-ABL kinase, inhibiting its activity and controlling cancer cell proliferation.
G-Protein Coupled Receptors (GPCRs): GPCRs are a large family of membrane proteins that interact with a variety of ligands, including hormones and neurotransmitters. The binding of ligands to GPCRs triggers intracellular signaling pathways that regulate many physiological processes. Drugs that target GPCRs, such as beta-blockers and antihistamines, are widely used to treat cardiovascular disease, allergies and other conditions.
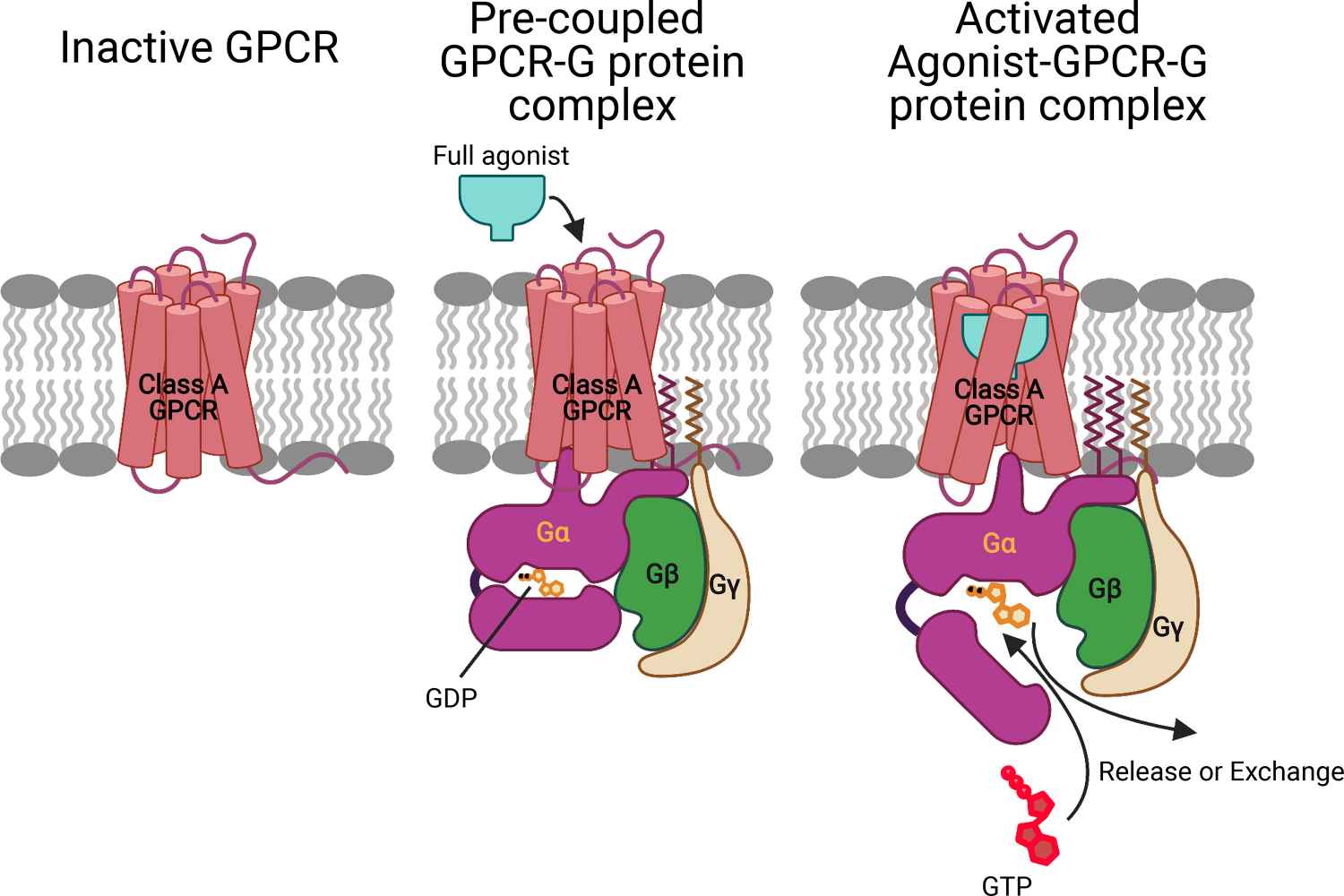 Figure 2. Mechanism for ligand activation of the GPCR–G protein complex. Prior to ligand binding, the inactive GP interacts with the inactive GPCR to open the intracellular region by breaking the TM3-TM6 tight link to form a stable pre-coupled complex. This pre-coupled complex remains at this resting state until an agonist binds to the GPCR-GP complex to open the intracellular region of GPCR and the tightly coupled Gα-GDP complex to form the fully activated agonist-GPCR-GP complex with the GDP available for exchange or release (Mafi et al., 2022).
Figure 2. Mechanism for ligand activation of the GPCR–G protein complex. Prior to ligand binding, the inactive GP interacts with the inactive GPCR to open the intracellular region by breaking the TM3-TM6 tight link to form a stable pre-coupled complex. This pre-coupled complex remains at this resting state until an agonist binds to the GPCR-GP complex to open the intracellular region of GPCR and the tightly coupled Gα-GDP complex to form the fully activated agonist-GPCR-GP complex with the GDP available for exchange or release (Mafi et al., 2022).
Creative Biostructure specializes in analyzing protein-ligand interactions, essential for drug discovery and understanding biological systems. If you're curious about how our services can improve your research on protein-ligand interactions, feel free to contact us for a custom quote.
Reference
- Acuner Ozbabacan, S. E., Engin, H. B., Gursoy, A., & Keskin, O. (2011). Transient protein-protein interactions. Protein Engineering Design and Selection, 24(9), 635–648.
- Du, X., Li, Y., Xia, Y.-L., Ai, S.-M., Liang, J., Sang, P., Ji, X.-L., & Liu, S.-Q. (2016). Insights into protein–ligand interactions: Mechanisms, models, and methods. International Journal of Molecular Sciences, 17(2), 144.
- Mafi, A., Kim, S.-K., & Goddard, W. A. (2022). The mechanism for ligand activation of the GPCR–G protein complex. Proceedings of the National Academy of Sciences, 119(18).
- Sharma, K., Balfanz, S., Baumann, A., & Korsching, S. (2018). Full rescue of an inactive olfactory receptor mutant by elimination of an allosteric ligand-gating site. Scientific Reports, 8(1), 9631.
