Structural Research of Oxygenases
Oxygenases are widely distributed in nature and participate in catalyzing the incorporation of molecular oxygen into various substrates, and the cofactors of the reaction include heme, non-heme iron, copper, and flavin. In recent years, it has been found that oxygenases play a crucial role in the metabolic transformation, synthesis, and degradation reactions of drugs and various natural compounds such as amino acids, lipids, hormones, and vitamins. It is significant to explore the potential functions of oxygenases, and the first step is to perform structural analysis.
Structural analysis of cyclooxygenase
The two mammalian cyclooxygenase isoforms, COX-1 and COX-2, both catalyze the incorporation of two oxygen atoms into arachidonic acid (AA) to produce prostaglandin G2 (PGG2), in which 15-hydroperoxides are reduced to form PGH2, which plays an important role in inflammatory responses. The structure of the di-peroxide-forming reaction of COX has been elucidated by electron paramagnetic resonance (EPR), kinetic analysis, X-ray crystallography, and mutagenesis experiments. The analyses showed that COX-1 and COX-2 form homodimers and that each COX monomer contains three structural domains, including an epidermal growth factor (EGF) domain, a membrane-bound domain, and a catalytic domain.
Advances in the structure of heme oxygenase (HO)
Heme oxygenase (HO) catalyzes the degradation of heme to biliverdin, carbon monoxide, and iron and regulates the levels of cytotoxic cofactors. As seen in the X-ray crystal structure, the two HO isomers, HO1 and HO2, share a catalytic core structure and both contain a C-terminal hydrophobic membrane anchor. Within the core is a pocket formed by two α-helices (proximal and distal helices). The proximal helix contains conserved histidine residues (His 25 in HO1 and His 45 in HO2), which are coordinated with the iron of the heme molecule. The distal helix contains conserved glycine residues, giving the helix a high degree of flexibility and enabling it to interact with bound heme.
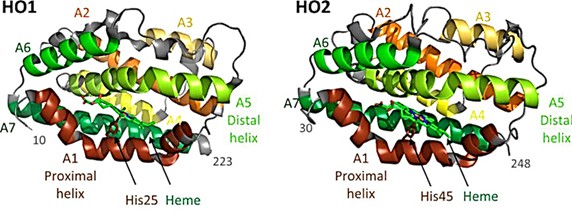 Figure 1. Structures of HO1 and HO2. (Kochert BA, et al., 2019)
Figure 1. Structures of HO1 and HO2. (Kochert BA, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Heme oxygenase-1-heme bound to NO | Rattus norvegicus | X-ray diffraction | 1.7 Å | 1J02 |
| Heme oxygenase 1 (HO-1) in complex with its substrate heme, crystal form B | Homo sapiens | X-ray diffraction | 2.58 Å | 1N3U |
| Heme oxygenase 1 (HO-1) in complex with its substrate heme | Homo sapiens | X-ray diffraction | 1.5 Å | 1N45 |
| Heme oxygenase (HO-1) in complex with heme binding dithiothreitol (DTT) | Rattus norvegicus | X-ray diffraction | 2.15 Å | 3I9T |
| Heme oxygenase-1 in complex with ferrous verdoheme | Rattus norvegicus | X-ray diffraction | 2.2 Å | 2ZVU |
| The ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of heme oxygenase-1 | Homo sapiens | X-ray diffraction | 2.59 Å | 1OYK |
| The NO-and CO-bound heme oxygenase | Neisseria meningitidis | X-ray diffraction | 2.1 Å | 1P3T |
| Heme oxygenase oxidatition of alpha- and gamma-meso-phenyl hemes | Homo sapiens | X-ray diffraction | 2.29 Å | 1S13 |
| Heme oxygenase in a complex with biliverdine | Homo sapiens | X-ray diffraction | 2.19 Å | 1S8C |
| HO-1 in complex with ferrous heme | Rattus norvegicus | X-ray diffraction | 2.3 Å | 1UBB |
| Xenon-bound heme-heme oxygenase-1 complex | Rattus norvegicus | X-ray diffraction | 1.9 Å | 1VGI |
| Heme oxygenase-. PCC6803 in complex with heme | Synechocystis sp. PCC 6803 | X-ray diffraction | 2.5 Å | 1WE1 |
| G139A, G139A-NO and G143H mutants of heme oxygenase-1 | Homo sapiens | X-ray diffraction | 1.88 Å | 1XJC |
| Bent-binding of cyanide to the heme iron in heme oxygenase-1 | Rattus norvegicus | X-ray diffraction | 1.85 Å | 2E7E |
| Heme oxygenase-1 in complex with 1-(adamantan-1-yl)-2-(1H-imidazol-1-yl) ethanone | Homo sapiens | X-ray diffraction | 1.54 Å | 3CZY |
| YcfD, a Ribosomal oxygenase | Escherichia coli K-12 | X-ray diffraction | 2.7 Å | 4LIU |
| Fe(II)/(alpha)ketoglutarate-dependent dioxygenase PrhA in complex with (alpha)ketoglutarate | Penicillium brasilianum | X-ray diffraction | 2.104 Å | 5YBN |
| Reduced IsdI in complex with heme | Staphylococcus aureus subsp. aureus N315 | X-ray diffraction | 1.88 Å | 3LGM |
| Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AusE | Aspergillus nidulans FGSC A4 | X-ray diffraction | 2.108 Å | 5YBL |
Table 1. Structural research of oxygenases.
Creative Biostructure has experts experienced in the structural analysis of membrane proteins. Our X-ray crystallographytechnology helps clients analyze the structure of oxygenases and further explore their potential functions in disease prevention and treatment.
We are also able to accurately analyze the structures of other biomolecules including, but not limited to, ribosomes, nucleic acids, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us for more information.
References
- Kochert BA, et al. Dynamic and structural differences between heme oxygenase-1 and -2 are due to differences in their C-terminal regions. J Biol Chem. 2019. 294(20):8259-8272.
- Mori T, Abe I. Structural basis for endoperoxide-forming oxygenases. Beilstein J Org Chem. 2022. 18:707-721.
