Structural Research of AAA Proteins
AAA proteins (ATPases Associated with diverse cellular Activities) utilize the energy of ATP hydrolysis to promote various functions in the cell, including dissolving protein aggregates, degrading proteins, or moving proteins across membranes. In general, these ATPases function primarily in the unfolding and disaggregation of proteins, but also in the extraction of proteins from membranes, two processes that are often combined with processing and degradation.
AAA proteins typically form homo-oligomers composed of six identical subunits assembled into a hexameric ring-like complex. Each subunit contains one or two highly conserved ATP-binding domains. Together they form a central pore-like channel lined with the pore loops, which plays a role in ATP hydrolysis-dependent substrate transport through this central pore.
Despite these common structural features, each AAA protein has unique structural characteristics that allow them to fulfill its specific biological purpose. For example, the AAA protein Msp1 in mitochondria is anchored to the mitochondrial outer membrane (OM) via its amino (N) -terminal TMH and assists to extract mistargeted tail-anchored proteins from the OM into the cytoplasm. In contrast to classical AAA proteins, Bcs1 specifically forms a homo-heptameric ring. This heptamer assembly of Bcs1 protein results in the formation of two large aqueous vestibules, one on the inner membrane vestibule, and a larger one on the matrix vestibule. The AAA domain and the amphipathic TMH delineate the inner membrane vestibule and matrix vestibule, respectively, with the intermediate domain forming a seal-like structure between the two. The structure of Bcs1 and the conformational differences between the apo and ADP states suggest an airlock-like mechanism for the translocation of Rieske protein Rip1. These structural studies benefit our understanding of how these specialized features ensure or contribute to the molecular mechanisms of specific AAA-ATPAse membrane translocators.
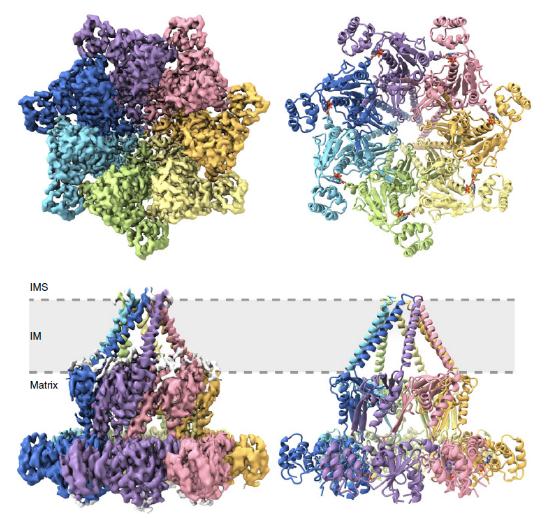 Figure 1. Top and side views of the Bcs1 structure in the ADP state. (Kater L, et al., 2020)
Figure 1. Top and side views of the Bcs1 structure in the ADP state. (Kater L, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| ADP state of the heptameric Bcs1 AAA-ATPase | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 3.40 Å | 6SH3 |
| Apo1 state of the heptameric Bcs1 AAA-ATPase | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 4.40 Å | 6SH4 |
| Apo2 state of the heptameric Bcs1 AAA-ATPase | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 4.60 Å | 6SH5 |
| Full-length heptameric Bcs1 AAA-ATPase (expressed in Komagataella pastoris) | Mus musculus | X-ray diffraction | 4.40 Å | 6UKO |
| Bcs1 AAA domain (expressed in Komagataella pastoris) | Mus musculus | X-ray diffraction | 2.17 Å | 6U1Y |
| Apo mBcs1 (expressed in Komagataella pastoris) | Mus musculus | Cryo-EM single particle analysis | 3.81 Å | 6UKP |
| ATPgammaS bound mBcs1 (expressed in Komagataella pastoris) | Mus musculus | Cryo-EM single particle analysis | 3.20 Å | 6UKS |
| Hexameric AAA protein Msp1 (E214Q)-substrate complex (expressed in E. coli) | Chaetomium thermophilum, E. coli | Cryo-EM single particle analysis | 3.50 Å | 6PE0 |
| Msp1-substrate complex in closed conformation (expressed in E. coli) | Chaetomium thermophilum, E. coli | Cryo-EM single particle analysis | 3.10 Å | 6PDW |
| Msp1-substrate complex in open conformation (expressed in E. coli) | Chaetomium thermophilum, E. coli | Cryo-EM single particle analysis | 3.70 Å | 6PDY |
Table 1. Structural research of AAA proteins.
In recent years, structural analysis of biomolecules using cryo-electron microscopy (cryo-EM) has become an important research direction in structural biology, especially for the study of membrane protein structures. Meanwhile, the obtained cryo-EM structures deepen the understanding of biomolecular functions, their roles in diseases, and targeted drug development.
Do you want to study your membrane protein structures using cryo-EM? If so, cryo-EM services from Creative Biostructure are your perfect solutions. We have been committed to structural biology and membrane protein research for a long time and have extensive experience in determining the structure of membrane proteins using single particle analysis (SPA). Our experienced experts will assist you with every step of the way, from sample preparation to data processing.
In addition to the structure determination of membrane proteins, we can accurately analyze biomolecules including but not limited to nucleic acids, small proteins, ribosomes, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please feel free to contact our experts, and they will offer professional and comprehensive solutions.
References
- Kater L, et al. Structure of the Bcs1 AAA-ATPase suggests an airlock-like translocation mechanism for folded proteins. Nature Structural & Molecular Biology. 2020. 27(2): 142-149.
- Tang W K, et al. Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein. Nature Structural & Molecular Biology. 2020. 27(2): 202-209.
- Wang L, et al. Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction. Elife. 2020. 9: e54031.
