Structural Research of Amt/Mep/Rh Channel Proteins
Amt/Mep/Rh channels are a family of membrane transporters that play a crucial role in the transportation of nitrogen-containing compounds in a variety of living organisms, including bacteria, fungi, and plants. Over the past few years, there have been significant advances in our understanding of the structural features and mechanisms of Amt/Mep/Rh channel proteins. One of the key breakthroughs in this area has been the elucidation of the crystal structures of several Amt/Mep/Rh channels.
For instance, the crystal structure of the ammonium transporter Amt-1 from Archaeoglobus fulgidus was recently solved, revealing a trimeric structure that enables the efficient passage of ammonium ions through a central channel. Subsequent studies have identified critical amino acid residues involved in substrate binding and transport, as well as regulatory domains that modulate the activity of these channels.
More recently, cryo-electron microscopy (cryo-EM) has emerged as a powerful tool for studying the structure and dynamics of Amt/Mep/Rh channels in their native membrane environment. Cryo-EM studies have provided high-resolution structural information on several Amt/Mep/Rh channels, including the eukaryotic Rh protein RhAG and the bacterial Mep-A transporter from Rhizobium leguminosarum.
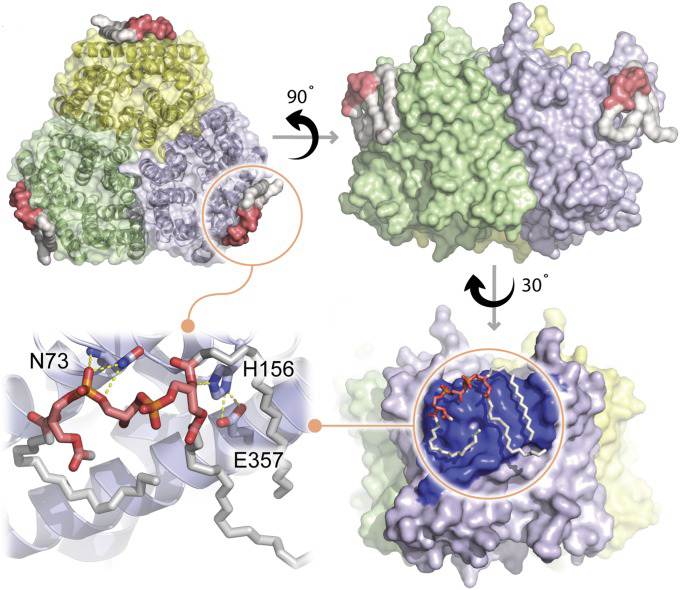 Figure 1. Crystal structure of AmtB Bound to TFCDL. (Patrick J W, et al., 2018)
Figure 1. Crystal structure of AmtB Bound to TFCDL. (Patrick J W, et al., 2018)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| AmtB ammonia channel (mutant) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.40 Å | 1U7G |
| AmtB ammonia channel (wild type) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.80 Å | 1XQF |
| AmtB ammonia channel (wild type) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.10 Å | 2NMR |
| AmtB ammonia channel in complex with GlnK (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.50 Å | 2NUU |
| AmtB ammonia channel in complex with inhibitory GlnK (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.96 Å | 2NS1 |
| AmtB ammonia channel in complex with phosphatidylglycerol (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.30 Å | 4NH2 |
| AmtB ammonia channel with bound TopFluor cardiolipin (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.45 Å | 6B21 |
| Amt-1 ammonium channel (expressed in E. coli) | Archaeoglobus fulgidus | X-ray diffraction | 1.72 Å | 2B2F |
| Rh protein, possible ammonia or CO2 channel (expressed in Methylococcus capsulatus) | Nitrosomonas europaea | X-ray diffraction | 1.85 Å | 3B9Y |
| Rh protein, possible ammonia or CO2 channel (expressed in E. coli) | Nitrosomonas europaea | X-ray diffraction | 1.30 Å | 3B9W |
| Human Rh C glycoprotein ammonia transporter (expressed in HEK293 cells) | Homo sapiens | X-ray diffraction | 2.10 Å | 3HD6 |
| Mep2 ammonium transceptor (expressed in Saccharomyces cerevisiae) | Saccharomyces cerevisiae | X-ray diffraction | 3.20 Å | 5AEX |
| Mep2 ammonium transceptor (R3) (expressed in Saccharomyces cerevisiae) | Candida albicans | X-ray diffraction | 1.47 Å | 5AEZ |
| Ammonium sensor/transducer (expressed in E. coli) | Candidatus Kuenenia stuttgartiensis | X-ray diffraction | 1.98 Å | 6EU6 |
Table 1. Structural Research of Amt/Mep/Rh Channel Proteins.
Creative Biostructure has extensive experience in the structural analysis of membrane proteins, including Amt/Mep/Rh channels. Our team of experts has decades of combined experience in the field of protein structure determination and analysis, and we have worked with clients from a wide range of industries and research fields.
We also offer additional services, such as protein-ligand binding studies, epitope mapping, and protein-protein interaction analysis. Our goal is to provide our clients with comprehensive, actionable insights into the structure and function of their proteins, enabling them to make informed decisions about drug discovery, biomolecular engineering, and other applications. If you are interested in our services, please feel free to contact us, and we will offer professional and comprehensive solutions.
References
- Patrick J W, et al. Allostery revealed within lipid binding events to membrane proteins. Proceedings of the National Academy of Sciences. 2018, 115(12): 2976-2981.
- Pflüger T, et al. Signaling ammonium across membranes through an ammonium sensor histidine kinase. Nature Communications. 2018, 9(1): 164.