Structural Research of Sorting Nexins
Sorting Nexins (SNXs) are proteins located in the cytoplasm, and several members of this family have been shown to have a role in promoting protein sorting. At the same time, the etiology of many diseases such as cancer, cardiovascular and neurodegenerative diseases is associated with dysregulation of sorting nexin function, suggesting an important role for SNXs in protein homeostasis. Structural analysis of Sorting Nexins can help scientists understand how Sorting Nexins interact with other proteins and reveal the mechanism of their role in cell membrane transport, endocytosis, and vesicle formation, which is important for understanding their functions. In addition, structural studies of Sorting Nexins can provide potential targets for drug design and treatment of related diseases.
A subgroup of classified linker proteins possesses a BAR structural domain at its C-terminus. The structure of the PDLIM7 PDZ structural domain bound to the C-terminus of SNX17 has been determined by NMR spectroscopy and reveals unconventional vertical peptide interactions. For example, the sorting linker protein SNX17 controls the recirculation of multiple transmembrane cargo proteins from the endosome to the cell surface. This requires association with a multisubunit commander transport complex that depends on the C-terminus of SNX17 via an unknown mechanism. SNX17 contains a type III PSD95/Dlg/Zo1 (PDZ) binding motif (PDZbm) that specifically binds the PDZ structural domain of PDLIM family proteins but not the other PDZ structural domains tested.
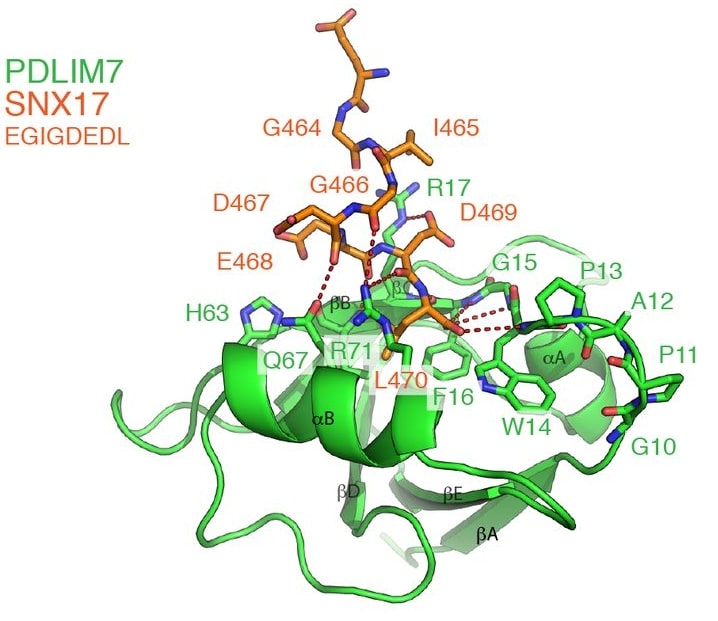 Figure 1. The structure of the PDLIM7 PDZ domain bound to the SNX17 C-terminus. (Healy MD, et al., 2022)
Figure 1. The structure of the PDLIM7 PDZ domain bound to the SNX17 C-terminus. (Healy MD, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Sorting Nexin 11 SNX11-PXe domain in dimer form: Homo sapiens E (expressed in E. coli) | Homo sapiens | X-ray diffraction | 3.50 Å | 6KOI |
| SNX11-PXe domain in complex with PI(3,5)P2 | Homo sapiens | X-ray diffraction | 2.14 Å | 6KOJ |
| SNX11/SNX10-PXe Chimera | Homo sapiens | X-ray diffraction | 2.00 Å | 6KOK |
Table 1. Structural research of Sorting Nexins.
A wide range of structural biology methods and techniques have been used by scientists in the structural studies of Sorting Nexins. Among them, X-ray crystallography is one of the most commonly used methods to reveal the three-dimensional structural information of Sorting Nexins by determining their high-resolution crystal structures. In addition, nuclear magnetic resonance (NMR) techniques have been used to study the structural and kinetic properties of Sorting Nexins. Methods such as cryo-electron microscopy (Cryo-EM) play an important role in studying the structure and intracellular localization of large complexes of Sorting Nexins.
Creative Biostructure is a technology company focused on the field of protein structural biology. By understanding the structure and function of proteins and their important role in cellular processes, it will provide scientists with valuable information and technical support. We can provide a variety of services related to Sorting Nexins structural research. Our services include crystal structure analysis of Sorting Nexins, NMR structure analysis, complex structure study, protein localization study, etc. Our team of professionals also has the experience and advanced equipment to provide high-quality structural biology services to a wide range of research institutions and pharmaceutical companies. For more information, please feel free to contact us for a more detailed introduction of our services.
References
- Worby CA, Dixon JE. Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol. 2002; 3 (12): 919-31.
- Hanley SE,Cooper KF. Sorting Nexins in Protein Homeostasis. Cells. 2020; 10 (1).
- Healy MD, et al. Proteomic identification and structural basis for the interaction between sorting nexin SNX17 and PDLIM family proteins. Structure. 2022; 30 (12): 1590-1602.e6.