Structural Research of Electron Transport Chain Complex I
The electron transport chain (ETC) is a fundamental process in aerobic respiration that produces ATP, the primary source of energy for living organisms. Among its key components is respiratory complex I, a large and intricate membrane protein complex responsible for the transfer of electrons from NADH to ubiquinone. In recent years, significant advances have been made in the structural research of complex I, which have contributed to our understanding of its biological function. For instance, the application of single-particle cryo-electron microscopy (cryo-EM) to study the catalytically active E. coli complex I, reconstituted into lipid nanodiscs, has revealed new information on the coupling mechanism of the enzyme. The structural analysis indicates a highly dynamic connection between the peripheral and membrane domains of the complex, with the peripheral domain assembly stabilized by unique terminal extensions and an insertion loop. Additionally, the membrane domain structure has unveiled novel dynamic features. Notably, the unconventional conformation of the conserved interface between the peripheral and membrane domains suggests an uncoupled state of the complex. The integration of these structural data has allowed for the proposal of a new simple hypothetical coupling mechanism for this molecular machine. Moreover, the determination of the peripheral arm structure at a resolution of 2.1 Å and that of the membrane domain at 3.7 Å has provided crucial insights into the structure and function of this complex, enhancing our knowledge of the respiratory chain in living systems.
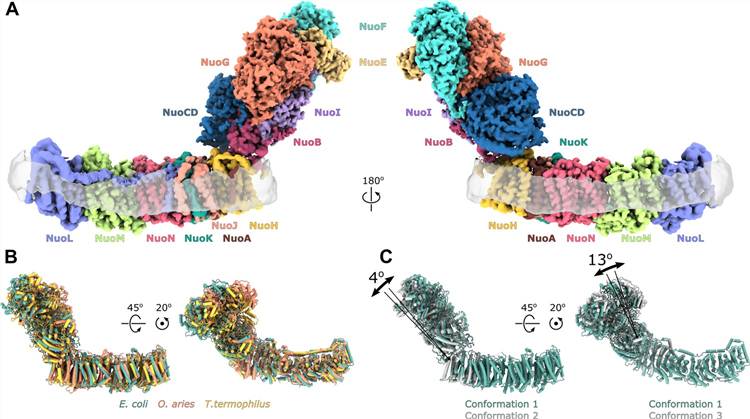 Figure 1. Architecture of E. coli respiratory complex I. (Kolata P, et al., 2021)
Figure 1. Architecture of E. coli respiratory complex I. (Kolata P, et al., 2021)
Table 1. Structural Research of Electron Transport Chain Complex I.
At Creative Biostructure, we specialize in analyzing the structure of membrane proteins (including electron transport chain complex I). Our team of highly skilled scientists utilizes various cutting-edge techniques such as cryo-EM, X-ray crystallography, and nuclear magnetic resonance (NMR) spectroscopy to obtain high-resolution images and detailed structural information. We provide a comprehensive range of services, including sample preparation, data collection and analysis, and structure determination, tailored to meet the specific needs of our clients. Our state-of-the-art facilities and equipment, combined with our extensive experience in membrane protein research, ensure timely and cost-effective delivery of high-quality results.
Whether you need high-resolution structures of electron transport chain complex I for research purposes or detailed structural information for drug development, we have the expertise and support to help you achieve your goals. Please feel free to contact us to learn more about our membrane protein structural analysis services. Our team of expert scientists is always available to address your inquiries and assist you in accomplishing your research objectives.
References
- Kolata P, Efremov R G. Structure of Escherichia coli respiratory complex I reconstituted into lipid nanodiscs reveals an uncoupled conformation. Elife. 2021, 10: e68710.
- Gutiérrez-Fernández J, et al. Key role of quinone in the mechanism of respiratory complex I. Nature Communications. 2020, 11(1): 4135.
- Bridges H R, et al. Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides. Science. 2023, 379(6630): 351-357.
