Structural Research of Hydrolases
Hydrolase is a general term for a type of enzyme that catalyzes hydrolysis reactions (such as trypsin, a type of hydrolysis enzyme that hydrolyzes polypeptide chains). Hydrolase can hydrolyze large molecule compounds into small molecule compounds, such as protein to amino acids, starch to glucose, etc. In recent years, with the help of X-ray crystallography, the research on the structure of hydrolase and its complex has made rapid progress.
The study of the structure of hydrolytic enzymes is crucial for elucidating the physiological processes of microorganisms in nature. At present, X-ray crystallography has provided unique insights into the mechanism of membrane phospholipid degradation mediated by hydrolases in Pseudomonas aeruginosa and other microorganisms. Scientific researchers used X-ray crystallography for the first time to identify the crystal structure of the membrane-bound phospholipase A1 (PlaF) from Pseudomonas aeruginosa under the resolution of 2.0 Å and successfully explained how PlaF degrades membrane phospholipids in a controlled way to allow bacteria to change their membrane composition to adapt to changing conditions.
The biosynthesis of bacterial cell walls has always been a focus of in-depth research. In 2021, the researchers showed the X-ray crystallography structure of the complex formed by the cell wall hydrolase SagB of Staphylococcus aureus and the membrane protein SpdC containing eight transmembrane helices at a resolution of 2.6 Å. The structure of the complex shows that the membrane protein provides a scaffold for the hydrolase to locate its active site to cut the glycan chain. For the first time, the analysis of the crystal structure of the complex assisted researchers to clarify the mechanism of bacteria releasing newly synthesized peptidoglycan chains from the membrane to complete the synthesis of mature peptidoglycan.
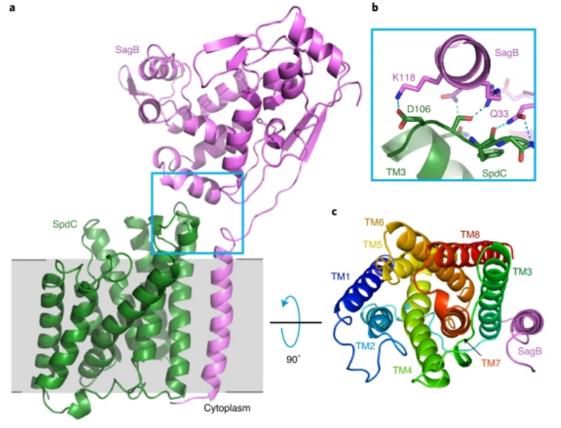 Figure 1. A crystal structure of the SagB-SpdC complex. (SCHAEFER K, et al., 2020)
Figure 1. A crystal structure of the SagB-SpdC complex. (SCHAEFER K, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Estrone Sulfatase | Homo sapiens | X-ray diffraction | 2.60 Å | 1P49 |
| Estrone Sulfatase (expressed in HEK293 cells) | Homo sapiens | X-ray diffraction | 2.04 Å | 8EG3 |
| Peptidoglycan release complex, SagB-SpdC | Staphylococcus aureus RF122, Staphylococcus aureus subsp. aureus NCTC 8325 | X-ray diffraction | 2.60 Å | 6U0O |
| Cryogenic human alkaline ceramidase 3 (ACER3) | Homo sapiens | X-ray diffraction | 2.60 Å | 6YXH |
| Fatty acid amide hydrolase | Rattus norvegicus | X-ray diffraction | 2.80 Å | 1MT5 |
| LpxH pyrophosphohydrase with bound lipid X & Mn2+ | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.65 Å | 5B49 |
| LpxH pyrophosphohydrase with bound lipid X & Mn2+ complex with lipid X, P21 form | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.72 Å | 5B4A |
| LpxH pyrophosphohydrase with bound lipid X & Mn2+ complex with lipid X, C2 form | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.60 Å | 5B4B |
| LpxH H10N mutant in complex with Mn2+ | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.96 Å | 5B4C |
| LpxH H10N mutant | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.75 Å | 5B4D |
| LpxI | Caulobacter vibrioides NA1000 | X-ray diffraction | 2.90 Å | 4GGM |
| LpxI D225A mutant | Caulobacter vibrioides NA1000 | X-ray diffraction | 2.52 Å | 4GGI |
| LpxK, the 4'-kinase of lipid A biosynthesis | Aquifex aeolicus VF5 | X-ray diffraction | 2.30 Å | 4EHW |
| Apo LpxK | Aquifex aeolicus VF5 | X-ray diffraction | 1.90 Å | 4EHX |
| ADP/Mg2+ LpxK | Aquifex aeolicus VF5 | X-ray diffraction | 2.20 Å | 4EHY |
| NAPE-PLD | Homo sapiens | X-ray diffraction | 2.65 Å | 4QN9 |
| PulA | Klebsiella oxytoca | X-ray diffraction | 2.88 Å | 2YOC |
| C2 domain of cytosolic phospholipase A2α bound to phosphatidylcholine | Gallus gallus | X-ray diffraction | 2.21 Å | 6IEJ |
| PlaF phospholipase A bacterial virulence factor | Pseudomonas aeruginosa | X-ray diffraction | 2.00 Å | 6I8W |
Table 1. Structural research of Hydrolases.
X-ray crystallography is a powerful technology widely used in structural biology. Its principle is to obtain protein structure images by irradiating X-ray beams and measuring crystal diffraction patterns. It is very suitable for analyzing the three-dimensional structure of membrane proteins. If accurate and comprehensive analysis and understanding of protein structure and function are required, Creative Biostructure's X-ray crystallography service is the gold standard choice. We use high-intensity X-ray and special detectors to provide high resolution protein crystal structure image.
References
- METZGER L E, et al. LpxI structures reveal how a lipid A precursor is synthesized. Nature Structural & Molecular Biology, 2012, 19(11): 1132–1138.
- OKADA C, et al. Crystal Structures of the UDP-diacylglucosamine pyrophosphohydrase lpxh from pseudomonas aeruginosa. Scientific Reports, 2016, 6(1).
- BRACEY M H, et al. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science, 2002, 298(5599): 1793–1796.
- HEALEY R D, et al. An automated platform for structural analysis of membrane proteins through serial crystallography. Cell Reports Methods, 2021, 1(6): 100102.
- SCHAEFER K, et al. Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization. Nature Microbiology, 2020, 6(1): 34–43.
- GHOSH D. Structure of human placental steroid sulfatase at 2.0 Angstrom resolution: Catalysis, quaternary association, and a secondary ligand site. The Journal of Steroid Biochemistry and Molecular Biology, 2023, 227: 106228.
- HERNANDEZ-GUZMAN F G, et al. Structure of human estrone sulfatase suggests functional roles of Membrane Association. Journal of Biological Chemistry, 2003, 278(25): 22989–22997.
- BLEFFERT F, et al. Structural, mechanistic, and physiological insights into phospholipase A-mediated membrane phospholipid degradation in pseudomonas aeruginosa. eLife, 2022, 11.
- HIRANO Y, et al. Structural basis of phosphatidylcholine recognition by the C2–domain of cytosolic phospholipase a2α. eLife, 2019, 8.
- EAST A, et al.Structural basis of Pullulanase membrane binding and secretion revealed by X-ray crystallography, molecular dynamics and biochemical analysis. Structure, 2016, 24(1): 92–104.
- MAGOTTI P, et al. Structure of human N -acylphosphatidylethanolamine-hydrolyzing phospholipase D: Regulation of fatty acid ethanolamide biosynthesis by bile acids. Structure, 2015, 23(3): 598–604.
- EMPTAGE R P, et al. Crystal structure of lpxk, the 4'-kinase of lipid a biosynthesis and atypical P-loop kinase functioning at the membrane interface. Proceedings of the National Academy of Sciences, 2012, 109(32): 12956–12961.