Structural Research of Vacuolar ATPase (V-ATPase)
Vesicular ATPase (V-ATPase) is a multisubunit protein that pumps protons into the inner lumen of organelles and undergoes ATP hydrolysis. V-ATPase controls intra- and extracellular pH and plays an essential role in the acidification of eukaryotes' intracellular vesicles, organelles, and extracellular environments. V-ATPase structure is found to be highly conserved in all eukaryotic cells. It comprises a cytoplasmic V1 complex for ATP hydrolysis and a membrane-embedded V0 complex for proton transfer.
Analysis of the structure and action mechanism of V-ATPase
In recent years, research has revealed the overall structure and function of V-ATPase. Hydrolysis of ATP in the hexameric A3B3 subcomplex in the V1 region leads to the cycling of the three pairs of A and B subunits between 'open', 'loose', and 'tight' conformations. This is followed by causing rotation of the rotor subcomplex. In yeast, the c ring contains eight c-subunits, one c′-subunit, and one c″-subunit, and the c ring surrounds the c″-subunit and the transmembrane α-helix of Voa1p. In mammals, the c-loop contains 9 c-subunits and 1 c″ subunit, and the c ring surrounds the transmembrane α-helices from the c″ subunit, the ATP6AP1/Ac45 subunit and the ATP6AP2/PRR subunit. In V0, the c ring rotates relative to the membrane-embedded C-terminal structural domain of subunit a, driving proton translocation across the membrane.
Structural research on the human V-ATPase
The researchers report cryo-electron microscopy structures of human V-ATPase in three rotational states with a resolution of up to 2.9 Å. With the aid of mass spectrometry analysis, protein subunits with associated N-linked glycans are constructed and glycolipids and phospholipids in the V0 complex are identified. The structure showed that the V1 complex consists of a top A3B3 head, a DF stem, three peripheral stems of the EG subunit, and a bottom collar formed by subunits H, C, and a-NTD from the V0 complex. The V0 complex consists of a c ring composed of c(1)-c(9) and c", the C-terminal structural domain adjacent to the subunit (a CTD), e and RNAseK, the ATP6AP1 and ATP6AP2 subunits within the c ring, and the d subunit located on the cytoplasmic side of the c ring connected to the central stalk of the V1 complex.
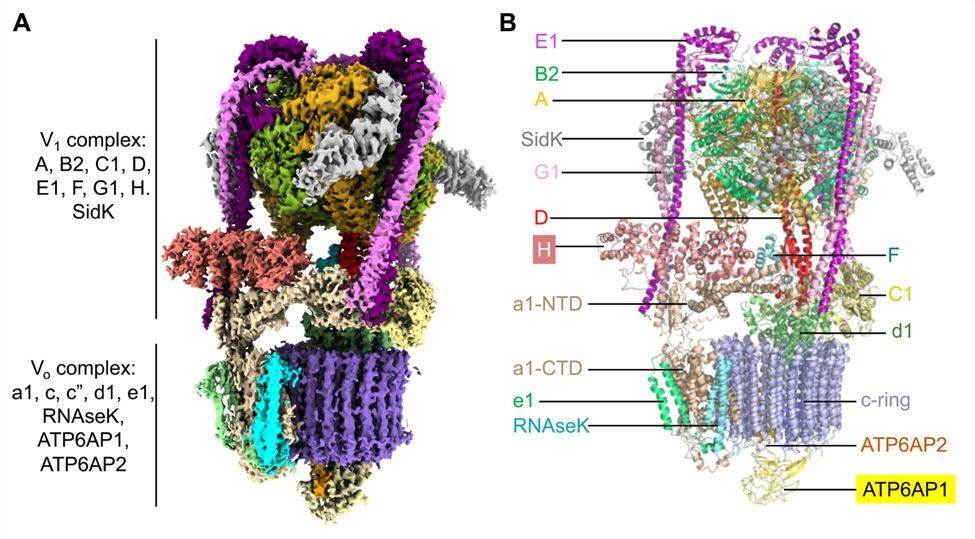 Figure 1. Cryo-EM structure of human V-ATPase. (Wang L, et al., 2020)
Figure 1. Cryo-EM structure of human V-ATPase. (Wang L, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Archazolid A bound to VO V-ATPase | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 2.8 Å | 7TAP |
| Bafilomycin A1 bound to VO V-ATPase | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 3.2 Å | 7TAO |
| Membrane-embedded motor of a eukaryotic V-ATPase | Saccharomyces cerevisiae S288C | Cryo-EM single particle analysis | 3.9 Å | 5TJ5 |
| V-ATPase Vph1-VO | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 3.2 Å | 6O7T |
| V-ATPase StV1-VO | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 3.1 Å | 6O7U |
| V-ATPase StV1-V1VO State 1 | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 6.6 Å | 6O7V |
| V-ATPase from bovine brain, state 1 | Bos taurus | Cryo-EM single particle analysis | 3.37 Å | 6XBW |
| V-ATPase from bovine brain, state 2 | Bos taurus | Cryo-EM single particle analysis | 3.79 Å | 6XBY |
| Vo motor in complex with 1 VopQ molecule | Saccharomyces cerevisiae S288C | Cryo-EM single particle analysis | 3.1 Å | 6PE4 |
| Vo motor in complex with 2 VopQ molecules | Saccharomyces cerevisiae S288C | Cryo-EM single particle analysis | 3.2 Å | 6PE5 |
| Lithium bound rotor ring of the V-ATPase | Enterococcus hirae | X-ray diffraction | 2.8 Å | 2CYD |
| Vo state3 prime | Saccharomyces cerevisiae S288C | Cryo-EM single particle analysis | 3.6 Å | 6M0S |
| Vo state3 | Saccharomyces cerevisiae S288C | Cryo-EM single particle analysis | 2.7 Å | 6M0R |
| V-ATPase state 1 | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 6.9 Å | 3J9T |
| V-ATPase state 2 | Saccharomyces cerevisiae | Cryo-EM single particle analysis | 7.6 Å | 3J9U |
| Porcine kidney V-ATPase with SidK, Rotary State 3 | Legionella pneumophila | Cryo-EM single particle analysis | 3.8 Å | 7U8R |
| Rotor ring with DCCD of the V- ATPase | Enterococcus hirae | X-ray diffraction | 2.4 Å | 2DB4 |
| The membrane rotor of the V-type ATPase | Enterococcus hirae | X-ray diffraction | 2.1 Å | 2BL2 |
| V-ATPase in state 1 with SidK and ADP | Homo sapiens | Cryo-EM single particle analysis | 3.1 Å | 6WM2 |
| V-ATPase in state 2 with SidK and ADP | Homo sapiens | Cryo-EM single particle analysis | 3.4 Å | 6WM3 |
| V-ATPase in state 3 with SidK and ADP | Homo sapiens | Cryo-EM single particle analysis | 3.6 Å | 6WM4 |
| The V1 region of V-ATPase in state 1 (focused refinement) | Homo sapiens | Cryo-EM single particle analysis | 2.9 Å | 6WLZ |
| The Vo region of V-ATPase in state 1 (focused refinement) | Homo sapiens | Cryo-EM single particle analysis | 3 Å | 6WLW |
| Subunit DF-assembly of the eukaryotic V-ATPase. | Saccharomyces cerevisiae S288C | X-ray diffraction | 3.18 Å | 4RND |
| Heterotrimeric EGChead peripheral stalk complex of the vacuolar ATPase | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.905 Å | 4DL0 |
| Transmembrane segment from subunit a from the V-ATPase | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | SOLUTION NMR | / | 2NVJ |
| A peptide derived from H+-V-ATPase subunit a | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | SOLUTION NMR | / | 2RPW |
Table 1. Structural research of the vacuolar ATPase (V-ATPase).
Creative Biostructure has a competent team of senior scientists and structural biologists. We have extensive experience in analyzing the complex structural properties of membrane proteins and are proficient in X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy. We have successfully determined the structures of numerous membrane proteins, including but not limited to V-ATPase.
Our structure analysis services are fully customized to clients' unique research requirements. For unrivaled expertise and all-encompassing solutions that are guaranteed to satisfy your structural biology requirements, please do not hesitate to contact us.
References
- Wang L, et al. Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly. Mol Cell. 2020. 80(3): 501-511. e3.
- Tan YZ, et al. Structure of V-ATPase from citrus fruit. Structure. 2022. 30(10): 1403-1410. e4.
- Indrawinata K, et al. Structural and functional understanding of disease-associated mutations in V-ATPase subunit a1 and other isoforms. Front Mol Neurosci. 2023. 16: 1135015.