Structural Research of Nodaviridae
The Nodaviridae are a group of enveloped RNA viruses divided into three subfamilies based on the host. Of these, alphanodavirus infects insects, betanodavirus infects fish, and gammanodavirus infects prawns. Research has found that Nodaviridae is a common cause of mortality in marine animals and insects, and although many vaccines have been developed against these viruses, their efficacy is relatively low. Therefore, more effective immunization strategies and comprehensive vaccines need to be developed. In recent years, a detailed understanding of the particle structure of Nodaviruses has provided valuable information for developing strategies to control outbreaks.
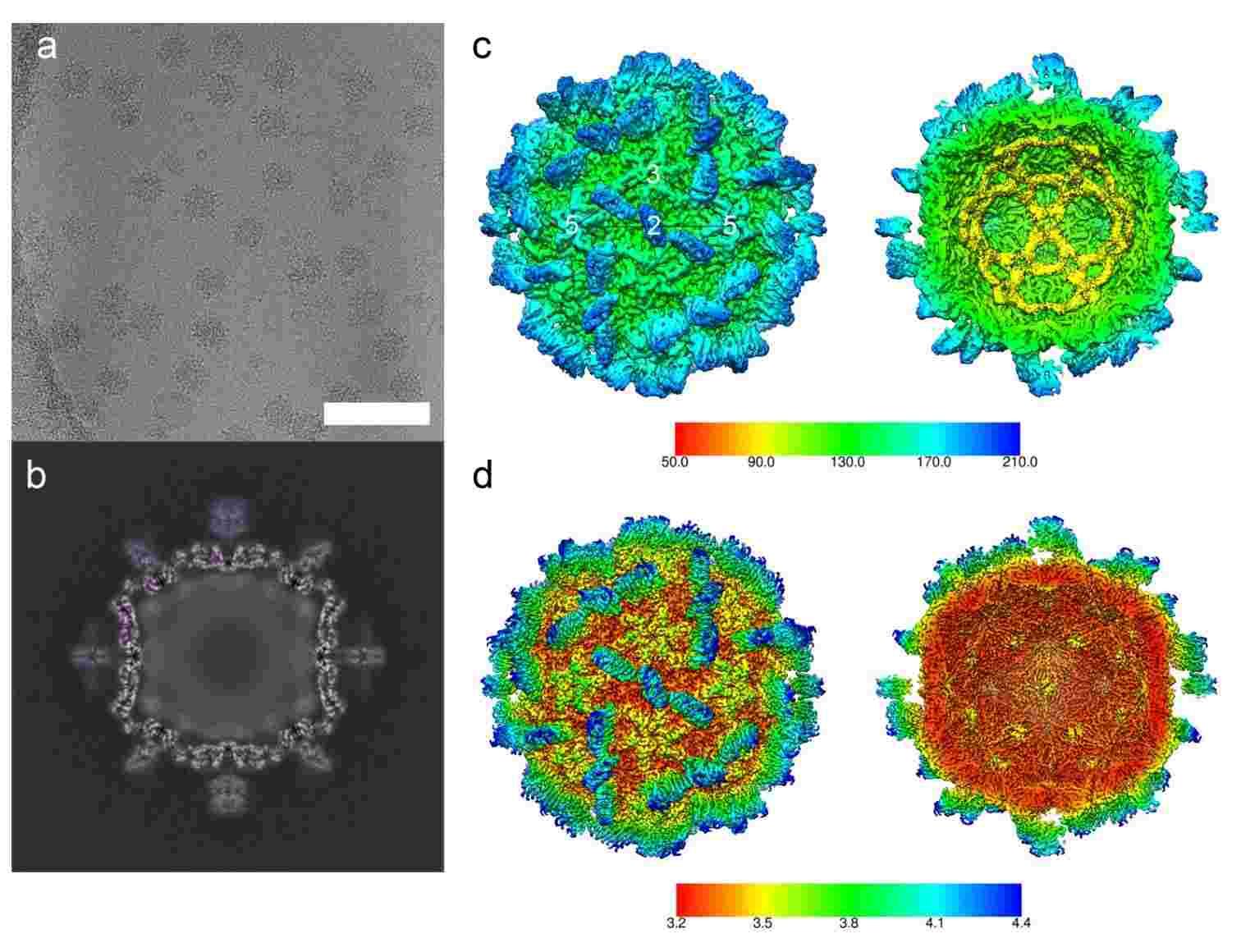 Figure 1. Cryo-electron microscopy and 3D reconstruction of MrNV VLPs. (Ho KL, et al, 2018)
Figure 1. Cryo-electron microscopy and 3D reconstruction of MrNV VLPs. (Ho KL, et al, 2018)
Genome Structure of Nodaviridae
The Nodaviridae genome contains two major infectious fragments, RNA1 and RNA2. During RNA replication RNA1 generates subgenomic RNA3. RNA1 (3.2 kb) encodes the RNA-dependent RNA polymerase (RdRp) and non-structural B2-like proteins and selects appropriate RNA templates and replication initiation sites to act. RNA2 (1.2 kb) encodes the viral capsid protein (CP) precursor protein α, which is important in the induction of apoptosis. Both RNA molecules are encapsidated in the same viral particle, capped at the 5' terminus and lacking the poly(A) tail at the 3' terminus. The N-terminal arginine-rich region of CP interacts with the RNA genome, and the C-terminal structural domain plays an essential role in host cell attachment and internalization.
Advances in Structural Research on Nodaviruses
Several three-dimensional structures of nodaviruses have been determined using X-ray crystallography and cryo-electron microscopy (cryo-EM). The structures show that the CPs of nodaviruses assemble into viral particles with an icosahedral structure and consist of a core jelly-roll topology that forms a face-to-face β-sandwich with two pairs of antiparallel β-sheet lamellae. In recent years, researchers have reconstructed the atomic resolution model of Macrobrachium rosenbergii nodavirus (MrNV) CP at 3.3 Å resolution using cryoEM. And cryoEM structures of purified Mr NV virion are obtained at 6.6 Å resolution. The data show that, unlike other known structures, Mr NV assembles a T = 3 capsid with a dimerization spike.
Creative Biostructure provides highly purified virus-like particles (VLPs) products for viral structure research. Viral structure analysis offers clients a detailed understanding of the three-dimensional (3D) arrangement of viral proteins. This information is invaluable for understanding viral entry mechanisms, antiviral drug and vaccine development, and the formulation of antiviral strategies.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V548 | Betanodavirus VLP (CP Proteins) | Betanodavirus | E. coli recombinant | CP (Capsid Protein) |
| CBS-V695 | Nervous necrosis viruses VLP (CP Protein) | Nervous necrosis virus | Yeast recombinant | Capsid protein |
| Explore All Nodaviridae Virus-like Particle Products | ||||
As a leading company in viral structure elucidation, Creative Biostructure offers meticulously engineered virus-like particles (VLPs) products that are powerful tools for a variety of applications in virus research and provide a safe alternative for researching viral entry mechanisms, immune responses, and vaccine development.
In addition, our team of scientists with extensive knowledge of virology and proficiency in cryo-electron microscopy (cryo-EM) helps clients resolve the 3D structure of virus particles and virus-like particles, which are crucial in drug discovery and vaccine development. Please feel free to contact us for more information. Choose us as your trusted partner to drive breakthroughs in virus research.
References
- Ho KL, et al. Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?. PLoS Biol. 2018. 16(10): e3000038.
- Shih TC, et al. Comprehensive Linear Epitope Prediction System for Host Specificity in Nodaviridae. Viruses. 2022. 14(7): 1357.
