Structural Research of Phosphatidic Acid Phosphatases
Phosphatidic acid phosphatase (PAP) plays an essential role in lipid homeostasis by controlling the cellular levels of substrate phosphatidic acid and product diacylglycerol, thereby ensuring lipid synthesis of triacylglycerol and membrane phospholipids. X-ray diffraction has been used to determine the three-dimensional structure of various PAPs, providing a detailed view of the action mechanisms, and helping to study the involvement in physiological activities such as lipid signal transduction, lipid droplet formation, vesicle transport, and phospholipid synthesis.
The role of PAP in lipid metabolism
PAP catalyzes the Mg2+-dependent dephosphorylation of phosphatidic acid to produce diacylglycerol (DAG), an important regulator of lipid homeostasis in eukaryotes. PAP-generated DAG is acylated to produce triacylglycerol (TAG) in endoplasmic reticulum membranes, which is then incorporated into lipid droplets. Due to its function in TAG synthesis, PAP could be developed as a drug target to ameliorate obesity or lipodystrophy.
Structural analysis of Lipin/Pah PAP
The Lipin/Pah PAP structure retains two regions, termed N-Lip and C-Lip. The crystal structure of PAP is refined to 3.0 Å resolution in the presence of calcium or magnesium. Phases are determined using single-wavelength anomalous diffraction from selenomethionine-rich proteins crystallized with calcium. The structure reveals an immunoglobulin-like structural domain (Ig-like structural domain, residues 20-139) and a catalytic structural domain of the haloalkanoate dehalogenase (HAD) superfamily (residues 140-321). The N-Lip is co-folded with the first 45 residues of the C-Lip to form the Ig-like structural domain, and the rest of the C-Lip forms the HAD-like catalytic structural domain.
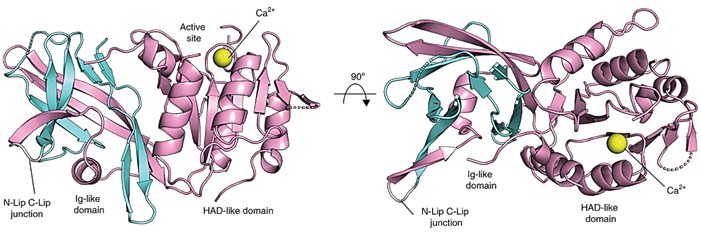 Figure 1. The general structure of Tt Pah2. (Khayyo VI, et al., 2020)
Figure 1. The general structure of Tt Pah2. (Khayyo VI, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Lipin-1 M-Lip domain | Mus musculus | X-ray diffraction | 1.467 Å | 7KIH |
| Lipin-2 M-Lip domain | Mus musculus | X-ray diffraction | 2.523 | 7KIQ |
| Lipin-1 M-Lip domain with zinc | Mus musculus | X-ray diffraction | 1.898 Å | 7KIL |
| Lipid phosphate phosphatase PgpB complex with PE | Escherichia coli K-12 | X-ray diffraction | 3.2 Å | 5JWY |
| An intramembrane CDP-DAG synthetase central (s200c/s258c, active mutant) | Thermotoga maritima MSB8 | X-ray diffraction | 3.4 Å | 4Q2E |
| Mitochondrial CDP-DAG synthase Tam41, delta 74 | Schizosaccharomyces pombe 972h- | X-ray diffraction | 2.261 Å | 6IG4 |
| Tam41 complex with CTP-Mg | Firmicutes bacterium CAG:884 | X-ray diffraction | 2.6 Å | 7ECD |
| The first transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 2.25 Å | 5JKI |
| Phosphatidic acid Transporter Ups1/Mdm35 Void of Bound Phospholipid | Saccharomyces cerevisiae S288C | X-ray diffraction | 2.9 Å | 5JQL |
| Lipin/Pah Phosphatidic Acid Phosphatase | Tetrahymena thermophila | X-ray diffraction | 3 Å | 6TZY |
| Lipin/Pah Phosphatidic Acid Phosphatase | Tetrahymena thermophila SB210 | X-ray diffraction | 1.95 Å | 7LHK |
| Lipin phosphatidic acid phosphatase with magnesium | Tetrahymena thermophila | X-ray diffraction | 3 Å | 6TZZ |
Table 1. Structural research of phosphatidic acid phosphatases.
Creative Biostructure has a long history of research in structural biology and membrane proteins. We use X-ray crystallography to analyze the structure of phosphatidic acid phosphatase to further explore its role and physiological effects in lipid metabolism.
We offer flexible and customized solutions to your needs, including full-service support, specific project-based work, and technology transfer. If you are interested in our services, please contact us and we will provide you with professional and comprehensive solutions.
References
- Khayyo VI, et al. Publisher Correction: Crystal structure of a Lipin/Pah phosphatidic acid phosphatase. Nat Commun. 2020. 11(1):1734.
- Carman GM, Han GS. Fat-regulating phosphatidic acid phosphatase: a review of its roles and regulation in lipid homeostasis. J Lipid Res. 2019. 60(1):2-6.