Structural Research of TMEM16 Family Proteins
TMEM16 proteins, also known as anoctamin, include Ca2+-activated ion channels and lipid scramblases. They are involved in a variety of cellular functions, including ion transport, phospholipid scrambling, and regulation of other membrane proteins. The first two members of the family, TMEM16A and TMEM16B, function as Ca2+-activated Cl- channels (CaCC). Since TMEM16 proteins have been implicated in a variety of human diseases, understanding the structure of TMEM16 proteins is essential for deciphering the molecular mechanisms of their activation gating, and regulation.
Advances in TMEM16 protein structure research
Functional TMEM16 proteins are dimers with a double-tubular structure in which separate permeable pores are present in each subunit. Each monomer has a different Ca2+ sensitivity or ion selectivity. In contrast to the originally predicted 8-transmembrane (TM) topology, TMEM16 monomers consist of 10 TM fragments preceded by a long N-terminal cytoplasmic structural domain (NCD) and followed by a short C-terminal extension of TM10.TM7 and TM8 do not fully cross the membrane, and together with TM6, they form two highly conserved Ca2+-binding sites.
Overall structural analysis of the TMEM16A channel
Researchers used X-rays to resolve TMEM16A as the first high-resolution structure of the TMEM16 family. The structure shows that TMEM16 is a dimer arranged in a two-lobed "butterfly" fold, with each subunit containing two Ca2+ binding sites and ten transmembrane (TM) helices. Each monomer has a hydrophilic transmembrane groove that provides a pathway for the lipid headgroup to move across the membrane. Both termini are structured and located on the cytoplasmic side of the membrane. The α-helix and β-chain of the amino-terminal domain are organized in a ferredoxin-like fold. The three alpha helices at the carboxyl terminus wrap around the N-terminal structural domain of the adjacent subunit.
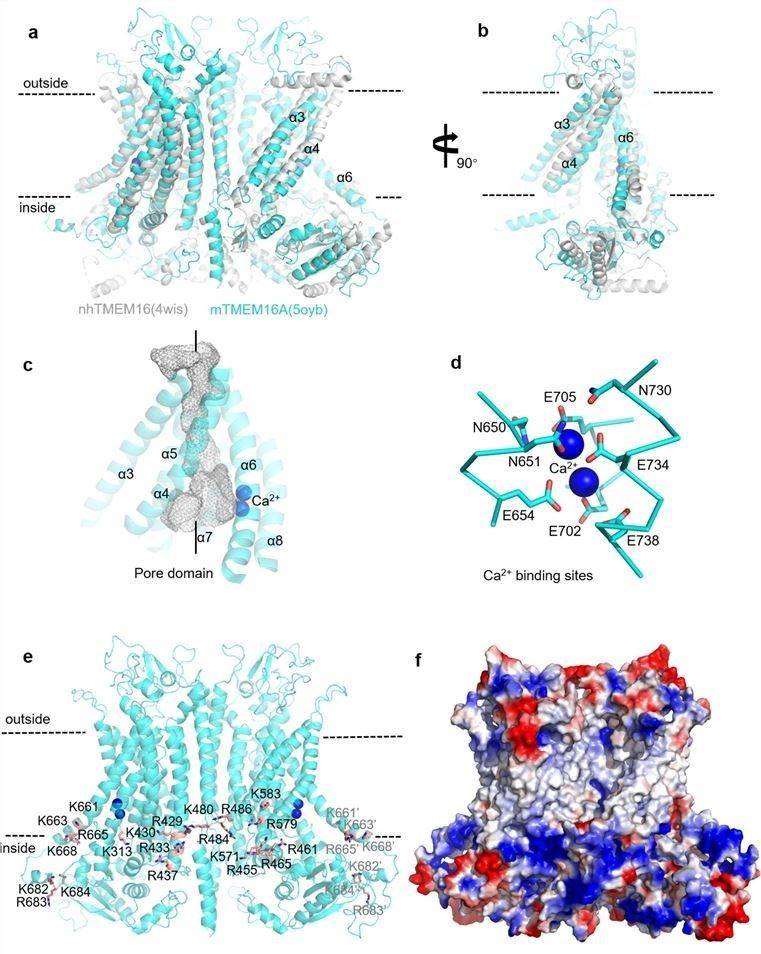 Figure 1. The molecular architecture of TMEM16A. (Shi S, et al., 2020)
Figure 1. The molecular architecture of TMEM16A. (Shi S, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Lipid scramblase nhTMEM16 in crystal form 1 | Fusarium vanettenii 77-13-4 | X-ray diffraction | 3.3 Å | 4WIS |
| TMEM16 lipid scramblase in crystal form 2 | Fusarium vanettenii 77-13-4 | X-ray diffraction | 3.4 Å | 4WIT |
| Calcium-bound nhTMEM16 lipid scramblase in nanodisc | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 3.7 Å | 6QMA |
| Calcium-bound nhTMEM16 lipid scramblase in nanodisc (closed state) | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 3.6 Å | 6QMB |
| Calcium-free nhTMEM16 lipid scramblase in nanodisc | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 3.8 Å | 6QM4 |
| Calcium-bound nhTMEM16 lipid scramblase in DDM | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 3.6 Å | 6QM5 |
| Calcium-free nhTMEM16 lipid scramblase in DDM | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 3.7 Å | 6QM6 |
| Calcium-bound nhTMEM16 lipid scramblase in nanodisc (open state) | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 3.6 Å | 6QM9 |
| afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ | Aspergillus fumigatus Af293 | Cryo-EM single particle analysis | 3.89 Å | 6DZ7 |
| afTMEM16 reconstituted in nanodiscs in the presence of Ca2+ | Aspergillus fumigatus Af293 | Cryo-EM single particle analysis | 4.05 Å | 6E0H |
| afTMEM16 reconstituted in nanodiscs in the presence of Ca2+ and ceramide 24:0 | Aspergillus fumigatus Af293 | Cryo-EM single particle analysis | 3.59 Å | 6E1O |
| nhTMEM16 L302A +Ca2+ in nanodiscs | Fusarium vanettenii 77-13-4 | Cryo-EM single particle analysis | 4 Å | 6OY3 |
| afTMEM16 in C14 lipid nanodiscs with MSP1E3 scaffold protein in the absence of Ca2+ | Aspergillus fumigatus A1163 | Cryo-EM single particle analysis | 3.3 Å | 7RX3 |
| afTMEM16 lipid scramblase in C18 lipid nanodiscs in the absence of Ca2+ | Aspergillus fumigatus A1163 | Cryo-EM single particle analysis | 3.07 Å | 7RXB |
| afTMEM16 in C18 lipid nanodiscs with MSP1E3 scaffold protein in the presence of Ca2+, full dimer | Aspergillus fumigatus A1163 | Cryo-EM single particle analysis | 2.28 Å | 7RXG |
| afTMEM16 in C22 lipid nanodiscs with MSP2N2 scaffold protein in the presence of Ca2+ | Aspergillus fumigatus Af293 | Cryo-EM single particle analysis | 3.5 Å | 7RWJ |
| afTMEM16 in C22 lipid nanodiscs with MSP1E3 scaffold protein in the presence of Ca2+ | Aspergillus fumigatus A1163 | Cryo-EM single particle analysis | 2.7 Å | 7RX2 |
| TMEM16K / Anoctamin 10 | Homo sapiens | X-ray diffraction | 3.2 Å | 5OC9 |
| TMEM16K / Anoctamin 10 (Form 2) | Homo sapiens | X-ray diffraction | 3.5 Å | 6R65 |
| TMEM16K / Anoctamin 10 in detergent (closed form) | Homo sapiens | Cryo-EM single particle analysis | 5.14 Å | 6R7Z |
| Calcium-free mTMEM16A chloride channel | Mus musculus | Cryo-EM single particle analysis | 4.06 Å | 5OYG |
| Calcium-bound mTMEM16A chloride channel | Mus musculus | Cryo-EM single particle analysis | 3.75 Å | 5OYB |
| TMEM16K / Anoctamin 10 in detergent (2mM Ca2+, closed form) | Homo sapiens | Cryo-EM single particle analysis | 3.47 Å | 6R7X |
| TMEM16K / Anoctamin 10 in detergent (low Ca2+, closed form) | Homo sapiens | Cryo-EM single particle analysis | 4.2 Å | 6R7Y |
| mTMEM16A ion channel | Mus musculus | Cryo-EM single particle analysis | 6.6 Å | 5NL2 |
| TMEM16A calcium-activated chloride channel in nanodisc | Mus musculus | Cryo-EM single particle analysis | 3.8 Å | 6BGI |
| Calcium-free mTMEM16A(ac)-I551A chloride channel | Mus musculus | Cryo-EM single particle analysis | 3.3 Å | 7B5D |
| 1PBC- and calcium-bound mTMEM16A(ac) chloride channel | Mus musculus | Cryo-EM single particle analysis | 2.85 Å | 7ZK3 |
| Calcium-free mTMEM16F lipid scramblase in digitonin | Mus musculus | Cryo-EM single particle analysis | 3.6 Å | 6QPB |
| mTMEM16F in lipid Nanodiscs in the presence of Ca2+ | Mus musculus | Cryo-EM single particle analysis | 2.94 Å | 8B8Q |
Table 1. Structural research of the TMEM16 family proteins.
Creative Biostructure is proud to offer a full suite of cutting-edge protein structural analysis services. Our cutting-edge technologies include as X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy, which allow us to delve deeper into the three-dimensional structure of proteins and provide valuable insights into their function and regulation.
Our experienced specialists are committed to providing timely and accurate results to our clients and advancing the development of protein structure analysis through collaboration with them. If you are interested in our services, please contact us for more details.
References
- Shi S, et al. Recent progress in structural studies on TMEM16A channel. Comput Struct Biotechnol J. 2020. 18: 714-722.
- Pedemonte N, Galietta LJ. Structure and function of TMEM16 proteins (anoctamins). Physiol Rev. 2014. 94(2): 419-459.
- Le SC, et al. Gating and Regulatory Mechanisms of TMEM16 Ion Channels and Scramblases.Front Physiol. 2021. 12: 787773.