Structural Research of Circoviridae
The family Circoviridae consists of viruses with a small circular single-stranded DNA (ssDNA) genome and includes two recognized genera, Circovirus and Cyclovirus, members of which are thought to cause fatal diseases in birds and pigs, and infections are characterized by damage to lymphoid tissue and immunosuppression. To understand the pathogenesis of economically important circovirus-associated diseases and develop related control strategies, it is essential to resolve the structure of circoviruses. Porcine circovirus (PCV) and beaked feather disease virus (BFDV) are among the more widely researched members of this family. PCV2 is the first circovirus capsid structure to be resolved at atomic resolution using X-ray crystallography. With the development of cryo-electron microscopy (cryo-EM) and NMR spectroscopy, researchers have resolved more structures of circoviruses at higher resolution.
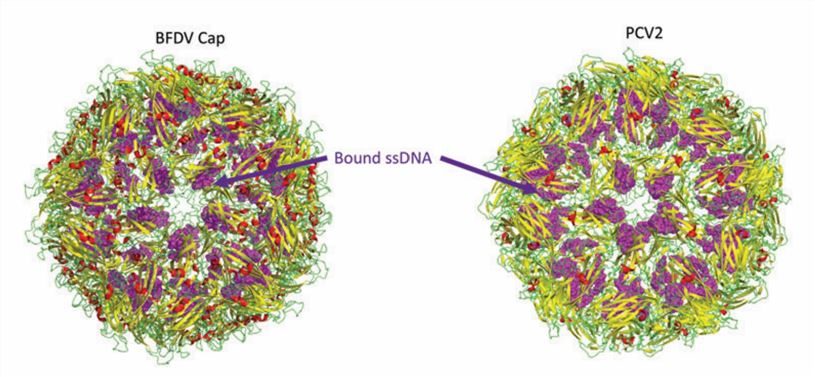 Figure 1. Structure of the circovirus BFDV and PCV2d VLP. (Nath BK, et al., 2021)
Figure 1. Structure of the circovirus BFDV and PCV2d VLP. (Nath BK, et al., 2021)
Genome Structure of Circoviruses
Members of the family Circoviridae are small, enveloped viruses with a circular, single-stranded DNA genome. The genome is approximately 2000 bp in length and contains two main open reading frames (ORFs), ORF1 and ORF2, encoding the Cap and Rep proteins, respectively. Cap is the structural protein of the virus and contains the major antigenic determinants with a highly conserved amino acid sequence. Rep is mainly involved in roll-over replication (RCR). Selective transcriptional splicing of ORF1 produces the Rep' protein, and the Rep-Rep' complex is required to facilitate viral replication. In addition to Rep and Cap, ORF3 of circoviruses encodes proteins involved in cell death during viral infection and plays an essential role in viral action mechanisms.
The Overall Structure of Circovirus
The circovirus has icosahedral and circular geometry, a diameter of approximately 20 nm, and T=1 symmetry. The capsid consists of 12 pentagonal flared pentamers (60 repeating subunits). The overall three-dimensional structure is conserved over large evolutionary distances in different species of circoviruses, although the sequences differ. The high-resolution atomic structure of PCV2 Cap shows that the Cap subunit includes a typical jelly roll structural domain. The jelly roll structural domain consists of two β-sheets consisting of four strands. The shorter loop connecting the β-strands modifies the surface of the capsid, while the longer loop mainly mediates interactions between the capsids. The N-terminal ARM region of PCV2 consists of an α-helix, which interacts with an arginine-rich NLS-B fragment near the proximal end of the β-barrel.
Creative Biostructure offers highly purified virus-like particles (VLPs) products for use in viral structure research. Our full range of products helps clients to understand the three-dimensional structure of the viral capsid and how it interacts with nucleic acids, which is of great application value in unraveling the mechanism of the viral life cycle, analyzing exogenous-mediated molecular delivery, antiviral drug, and vaccine development.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V716 | Porcine circoviruses VLP (Cap Protein) | Porcine circovirus | E. coli recombinant | Cap |
| CBS-V717 | Porcine circovirus 2 VLP (Cap Protein) | Porcine circovirus | E. coli recombinant | Cap protein |
| CBS-V718 | Porcine circovirus type 2 VLP (Capsid Protein) | Porcine circovirus | Yeast recombinant | Cap (Capsid protein) |
| Explore All Circoviridae Virus-like Particle Products | ||||
Creative Biostructure's scientists have a deep understanding of the structural resolution of viral particles and VLPs based on knowledge of viral structural biology and long-term project experience. We have cutting-edge instrumentation to characterize nanoscale VLP by accurately determining viral morphological features and confirming particle integrity.
In addition, our custom VLP construction services can satisfy specific requirements for formulating and modifying target VLPs. We are committed to working closely with clients to provide a tailored structure resolution strategy for VLP of interest. Please feel free to contact us for a detailed quote.
References
- Nath BK, et al. Structural Perspectives of Beak and Feather Disease Virus and Porcine Circovirus Proteins. Viral Immunol. 2021. 34(1): 49-59.
- Breitbart M, et al. ICTV Virus Taxonomy Profile: Circoviridae. J Gen Virol. 2017. 98(8): 1997-1998.
- Zhang Y, et al. Apoptosis Triggered by ORF3 Proteins of the Circoviridae Family. Front Cell Infect Microbiol. 2021. 10: 609071.