Structural Research of APC Superfamily
The amino acid/polyamine/organocation (APC) superfamily encompasses a diverse group of transmembrane transporters involved in the transport of small organic molecules such as amino acids, polyamines, and ions across the lipid bilayer. This superfamily is crucial for maintaining cellular homeostasis and is involved in many physiological processes, including nutrient uptake, ion homeostasis, and signaling.
Crystallography and cryo-electron microscopy have been used to determine the three-dimensional structures of various APC transporters, providing a detailed view of the transport mechanism. NKCC1 protein is a cation-chloride cotransporter involved in the transport of ions across cell membranes. The molecular structure of the protein has been determined, including its transmembrane domains and regulatory regions, and how it interacts with ions and other molecules. NKCC1 can transport ions across membranes through various mechanisms, including through the use of sodium-potassium pumps and the action of various transport proteins and channels. These studies suggest that a better understanding of the structure and mechanism of NKCC1 may lead to the development of new therapies for conditions such as hypertension and epilepsy, which are associated with the dysregulation of ion transport.
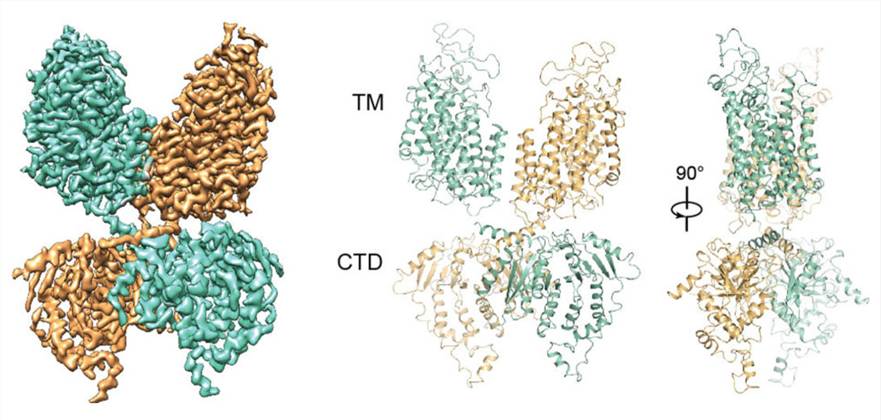 Figure 1. Overall structure of NKCC1 dimer. (Chew T A, et al., 2019)
Figure 1. Overall structure of NKCC1 dimer. (Chew T A, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Arginine Agmatine Antiporter (AdiC) (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.61 Å | 3LRB |
| AdiC (N22A, L123W mutant) with bound Arginine (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.00 Å | 3L1L |
| AdiC (N101A) in the open-to-out Arg+ bound conformation (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.00 Å | 3OB6 |
| AdiC in complex with agmatine (expressed in E. coli) | Escherichia coli | X-ray diffraction | 2.59 Å | 5J4N |
| AdiC, outward open substrate-free state (expressed in E. coli) | Escherichia coli | X-ray diffraction | 1.69 Å | 7O82 |
| AdiC in complex with a Fab fragment (expressed in E. coli) | Salmonella enterica | X-ray diffraction | 3.20 Å | 3NCY |
| ApcT Transporter (expressed in E. coli) | Methanocaldococcus jannaschii | X-ray diffraction | 2.32 Å | 3GIA |
| Glutamate-GABA antiporter GadC (expressed in E. coli) | Escherichia coli | X-ray diffraction | 3.10 Å | 4DJK |
| AgcS sodium/alanine symporter with bound L-alanine (expressed in E. coli) | Methanococcus maripaludis | X-ray diffraction | 3.24 Å | 6CSE |
| BasC alanine-serine-cysteine exchanger, open-inward conformation (expressed in E. coli) | Carnobacterium sp. AT7 | X-ray diffraction | 2.92 Å | 6F2G |
| NKCC1 TM domain (expressed in Spodoptera frugiperda) | Danio rerio | Cryo-EM single particle analysis | 2.90 Å | 6NPH |
| NKCC1 cation-chloride cotransporter (CCC), partially loaded, inward open state (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.46 Å | 6PZT |
| KCC1 cation-chloride cotransporter (CCC) in glyo-diosgenin (GDN) detergent, 150 mM KCl (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.90 Å | 6KKR |
| KCC1 Δ19 cation-chloride cotransporter (CCC) in NaCl (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.12 Å | 7AIP |
| KCC1 cation-chloride cotransporter (CCC) with bound VU0463271, outward-open state (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.50 Å | 7TTI |
| KCC3 cation-chloride cotransporter (CCC) (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.60 Å | 7D90 |
| KCC3b cation-chloride cotransporter (CCC), wild-type in NaCl (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.64 Å | 7NGB |
| KCC4 cation-chloride cotransporter (CCC) (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.90 Å | 7D99 |
| KCC4 cation-chloride cotransporter (CCC) in nanodiscs (expressed in Sf9 cells) | Mus musculus | Cryo-EM single particle analysis | 3.65 Å | 6UKN |
| KimA K+/H+ symporter, inward-facing occluded state (expressed in E. coli) | Bacillus subtilis | Cryo-EM single particle analysis | 3.70 Å | 6S3K |
Table 1. Structural Research of Amino Acid/Polyamine/Organocation (APC) Superfamily.
At Creative Biostructure, we understand the importance of obtaining high-quality, reliable structural information for the study of protein function and drug discovery. Our team of experienced scientists, state-of-the-art equipment, and proven track record of success make us the ideal partner for your structural biology needs. We have a team of experienced scientists who have a deep understanding of the complexities involved in these processes and can offer expert guidance to ensure the success of your project. If you are interested in learning more about our services and how we can help you achieve your goals, please do not hesitate to contact us.
References
- Chew T A, et al. Structure and mechanism of the cation-chloride cotransporter NKCC1. Nature. 2019, 572(7770): 488-492.
- Zhao Y, et al. Structure of the human cation-chloride cotransport KCC1 in an outward-open state. Proceedings of the National Academy of Sciences. 2022, 119(27): e2109083119.