Structural Research of Picornaviridae
Viruses classified as Picornaviridae have a single-stranded, highly diverse positive-stranded RNA genome that displays an envelope-less small icosahedral structure. All members of the family are characterized by three capsid proteins (CP) with β-barrel folds and RNA-directed replication with YGDD sequence motifs. Picornaviruses infect a wide range of animal and human hosts, ranging from mild febrile illnesses to severe cardiac, hepatic, and central nervous system diseases. Among them, Poliovirus is the typical research subject of the family and is the causative agent of poliovirus. Structural research on polioviruses is essential to support global poliovirus eradication efforts.
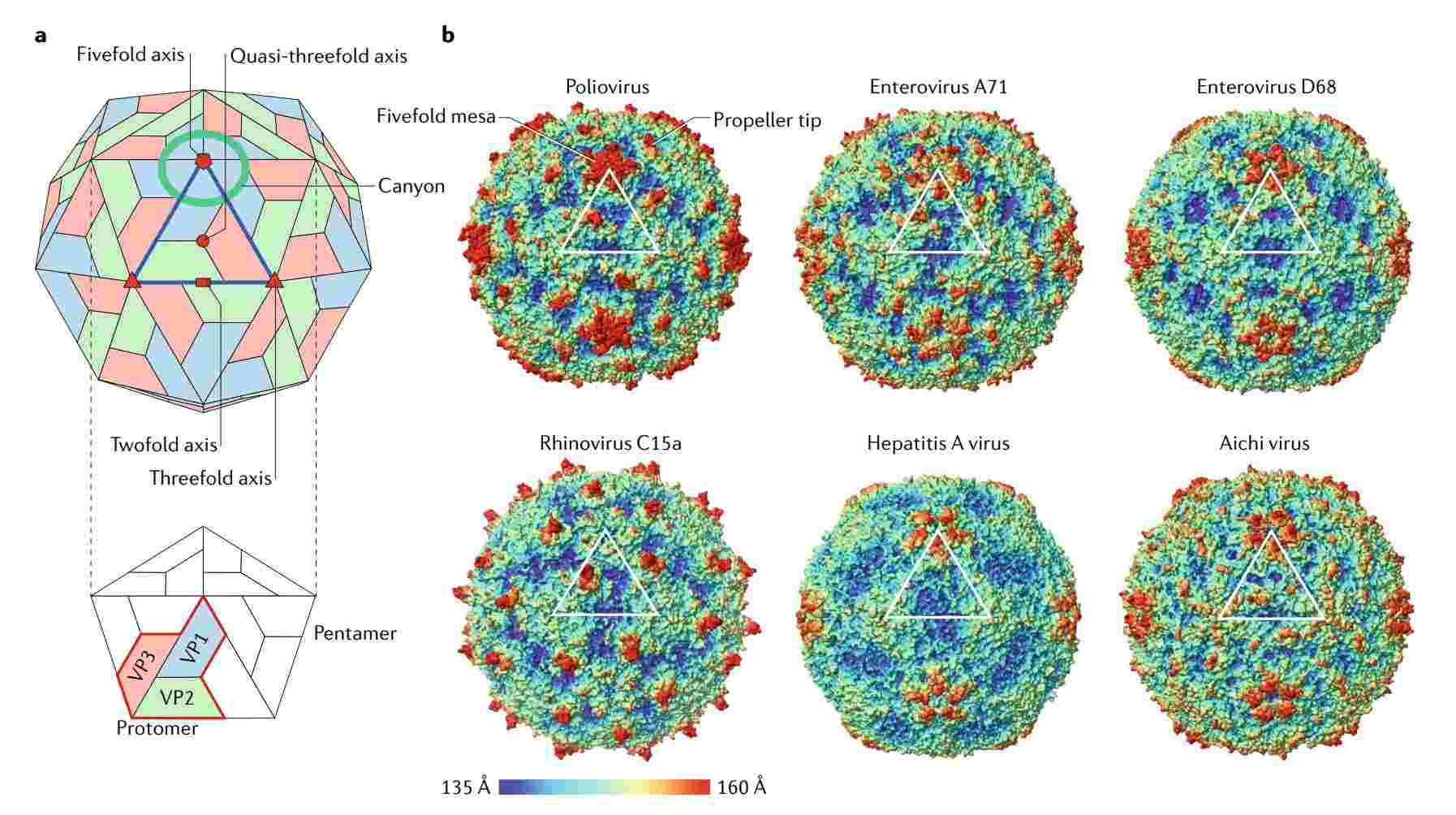 Figure 1. Picornavirus structure. (Baggen J, et al, 2018)
Figure 1. Picornavirus structure. (Baggen J, et al, 2018)
Morphological Features of Picornaviruses Viral Particles
Viral particles do not have an envelope, and the capsid surrounds the ssRNA core, which is 30-32nm in diameter. The capsid consists of 60 identical units. Proteins 1B, 1C, 1D, and uncut 1AB all have core structures containing eight-stranded β-sandwiches ('β-barrels'), which are stacked together in the capsid with T=1, pseudo-T=3, and icosahedral symmetry. Members of different genera differ in the external loops connecting the β-strands. Viral particles are assembled via pentameric intermediates, with proteins within each pentamer linked together by the N-termini of the three major CPs, with the C-termini located on the outer surface of the capsid. Research has revealed that CP sequences in many genera may contain conserved "inhibitor binding sites" that can bind active antiviral drugs.
Advances in Structural Research on Picornaviruses
Poliovirus, the best-known enterovirus, was the first animal RNA virus for which the complete genome sequence was determined and the three-dimensional structure resolved by X-ray crystallography. In recent years, researchers have also conducted in-depth structural studies of non-polio enteroviruses (NPEVs) using cryo-electron microscopy (cryo-EM) at 3 Å resolution, and several NPEVs have become serious public health threats. The understanding of the structural diversity of the small RNA virus family has been enhanced by developments in structural biology (high-resolution cryo-electron microscopy (cryo-EM) and high-speed fixed-target X-ray crystallography at X-ray free-electron lasers). Mechanisms of viral genome replication and assembly are explained and potential targets for antiviral therapy are described, contributing to developing antiviral therapies and vaccines.
It contributes to the development of antiviral therapies and vaccines by researching the virion structure, mechanisms of viral particle assembly and release, host-virus interactions, and potential targets for antiviral therapy. For viral structure elucidation, we offer a comprehensive range of high-quality virus-like particles (VLPs) products that closely mimic the morphology and surface features of target viruses, providing a safe and controlled system for researching viral structure without infectious genetic material.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V566 | Coxsackievirus B3 VLP (VP1; VP2; VP3; VP4 Proteins) | Coxsackievirus | Insect cell recombinant | VP1; VP2; VP3; VP4 |
| CBS-V585 | Enterovirus 71 VLP (P1 (VP0; VP1; VP3); 3CD Proteins) | Enterovirus 71 | Yeast recombinant | P1 (VP0; VP1; VP3); 3CD |
| CBS-V722 | Poliovirus -1 VLP (Capsid Protein) | Poliovirus | Yeast recombinant | Capsid |
| Explore All Picornaviridae Virus-like Particle Products | ||||
Creative Biostructure is a leader in viral structural biology, offering meticulously engineered virus-like particles (VLPs) that are invaluable tools for a wide range of applications, including vaccine development, drug discovery, and diagnostic analysis.
Our team of scientists specializes in viral structural analysis, utilizing advanced techniques including cryo-electron microscopy (cryo-EM), X-ray crystallography, and other cutting-edge imaging systems to reveal the complex structure of viruses. With expertise and precision, we provide comprehensive insights into viral proteins and the entire viral assembly. Please feel free to contact us with your requirements for viral structure analysis.
References
- Baggen J, et al. The life cycle of non-polio enteroviruses and how to target it. Nat Rev Microbiol. 2018. 16(6): 368-381.
- Zell R, et al. ICTV Virus Taxonomy Profile: Picornaviridae. J Gen Virol. 2017. 98(10): 2421-2422.
- Andino R, et al. The Picornaviridae Family: Knowledge Gaps, Animal Models, Countermeasures, and Prototype Pathogens. J Infect Dis. 2023. 228(Suppl 6): S427-S445.
