Structural Research of Oxidases
Oxidase is a type of enzyme that catalyzes oxidation reactions. Its primary function is to react the substrate (substrate) with oxygen to generate products such as water, carbon dioxide, and hydrogen peroxide, and release energy. Oxidases are widely present in organisms and participate in various life activities such as physiological metabolism, cell signal transduction, and waste disposal.
Single crystal X-ray diffraction can reveal the oxidase's molecular structure and composition and the enzyme mechanism's critical properties. Specifically, this technology can help scientists understand the three-dimensional structure of oxidase, including the precise location and coordination of amino acid residues, dehydrogenase, and other functional regions in proteins. In addition, single-crystal X-ray diffraction technology can further study the interaction between oxidase and substrates and ligands, and understand the essential characteristics of the oxidase catalytic reaction mechanism. This structural information has important guiding significance for developing efficient drugs and new catalysts and can provide beneficial support for research in disciplines such as biochemistry, biotechnology, and nanotechnology.
Monoamine oxidase B (MAO-B) is a proven drug target for Parkinson's disease. Chromone derivatives are a potential and reversible MAO-B inhibitor. To study the inhibitory mechanism of chromone derivatives on MAO-B, scientists determined the crystal structure of the human MAO-B complex with three chromone analogues with different substituents on the outer aromatic ring at 1.6-1.8Å resolution, indicating that they are all bound in the cavity of the active site of the protein. The chromone part is located in front of the FAD cofactor.
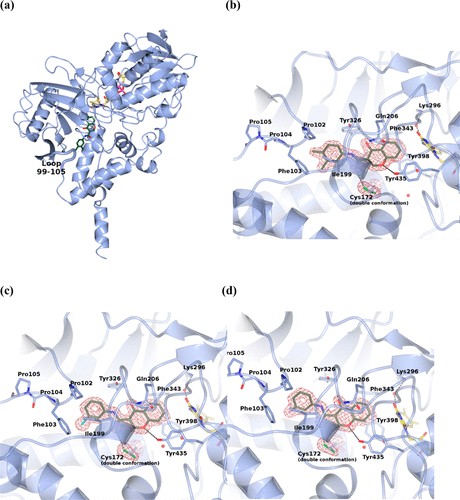 Figure 1. Crystal structure of human MAO-B in complex with chromone inhibitors. (REIS, et al., 2019)
Figure 1. Crystal structure of human MAO-B in complex with chromone inhibitors. (REIS, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Monoamine Oxidase B | Homo sapiens | X-ray diffraction | 3.00 Å | 1GOS |
| Monoamine Oxidase B with bound Isatin | Homo sapiens | X-ray diffraction | 1.70 Å | 1OJA |
| Monoamine Oxidase B with bound Tranylcypromine | Homo sapiens | X-ray diffraction | 2.20 Å | 1OJB |
| Monoamine Oxidase B with bound N-(2-aminoethyl)-p-chlorobenamide | Homo sapiens | X-ray diffraction | 2.40 Å | 1OJC |
| Monoamine Oxidase B with bound Lauryldimethyl-amine N-Oxide | Homo sapiens | X-ray diffraction | 3.10 Å | 1OJD |
| Monoamine Oxidase B with bound 1.4-Diphenyl-2-butene | Homo sapiens | X-ray diffraction | 2.30 Å | 1OJ9 |
| Monoamine Oxidase A | Rattus norvegicus | X-ray diffraction | 3.20 Å | 1O5W |
| Monoamine Oxidase A with bound Clorglycine | Homo sapiens | X-ray diffraction | 3.00 Å | 2BXR |
| Monoamine Oxidase A crystal form B | Homo sapiens | X-ray diffraction | 3.15 Å | 2BXS |
| Monoamine Oxidase B with bound Deprenyl | Homo sapiens | X-ray diffraction | 2.20 Å | 2BYB |
| Monoamine Oxidase A with bound Harmine | Homo sapiens | X-ray diffraction | 2.20 Å | 2Z5X |
| Monoamine Oxidase G110A mutant with bound Harmine | Homo sapiens | X-ray diffraction | 2.17 Å | 2Z5Y |
| Alternative oxidase (AOX), cyanide-insensitive | Trypanosoma brucei brucei | X-ray diffraction | 2.85 Å | 3VV9 |
| AOX with ascofuranone derivative | Trypanosoma brucei brucei | X-ray diffraction | 2.59 Å | 3VVA |
| AOX with colletochlorin B | Trypanosoma brucei brucei | X-ray diffraction | 2.30 Å | 3W54 |
Table 1. Structural research of oxidases.
Creative Biostructure is dedicated to providing X-ray crystallography-based membrane protein analysis services. Our advanced, professional, stable, accurate and reliable technology can help our clients better understand the structure, function and corresponding metabolic pathways of membrane proteins. Our services include a full range of support from crystal growth to structural analysis, which can provide our clients with high-quality data and support to obtain more research results. The professionalism and enthusiasm of our team have earned us high acclaim and customer trust in the field. By choosing our services, you will receive high-quality membrane protein crystal analysis services and complete post-process data analysis, interpretation and reporting.
References
- SHIBA T, et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proceedings of the National Academy of Sciences, 2013, 110(12): 4580–4585.
- SON S-Y, et al. Structure of human monoamine oxidase A at 2.2-Å resolution: The control of opening the entry for substrates/inhibitors. Proceedings of the National Academy of Sciences, 2008, 105(15): 5739–5744.
- DE COLIBUS L, et al. Three-dimensional structure of human monoamine oxidase A (Mao a): Relation to the structures of rat Mao A and human Mao B. Proceedings of the National Academy of Sciences, 2005, 102(36): 12684–12689.
- MA J, et al. Structure of rat monoamine oxidase a and its specific recognitions for substrates and inhibitors. Journal of Molecular Biology, 2004, 338(1): 103–114.
- BINDA C, et al. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. Proceedings of the National Academy of Sciences, 2003, 100(17): 9750–9755.
- BINDA C, et al. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nature Structural Biology, 2001, 9(1): 22–26.
- REIS J, et al. Tight-binding inhibition of human monoamine oxidase B by Chromone analogs: A Kinetic, crystallographic, and biological analysis. Journal of Medicinal Chemistry, 2018, 61(9): 4203–4212.