Structural Research of Secoviridae
Members of the Secoviridae family are enveloped viruses with single- or two-part (RNA-1 and RNA-2) linearly positive-sense ssRNA genomes. Most of the known members infect dicotyledonous plants, and many are essential plant pathogens, such as cowpea mosaic virus (CPMV) and apple latent spherical virus (ALSV). One of the major reasons why Secoviruses pose a significant agro-economic hazard is that they can encode specific proteins that allow them to move through the plant and counteract plant defense mechanisms. Research on the structure of Secoviruses is essential for understanding the mechanisms of virus infestation and developing control strategies. Meanwhile, a deeper understanding of the structure and protein function of Secoviruses can provide a basis for research and application in the life sciences.
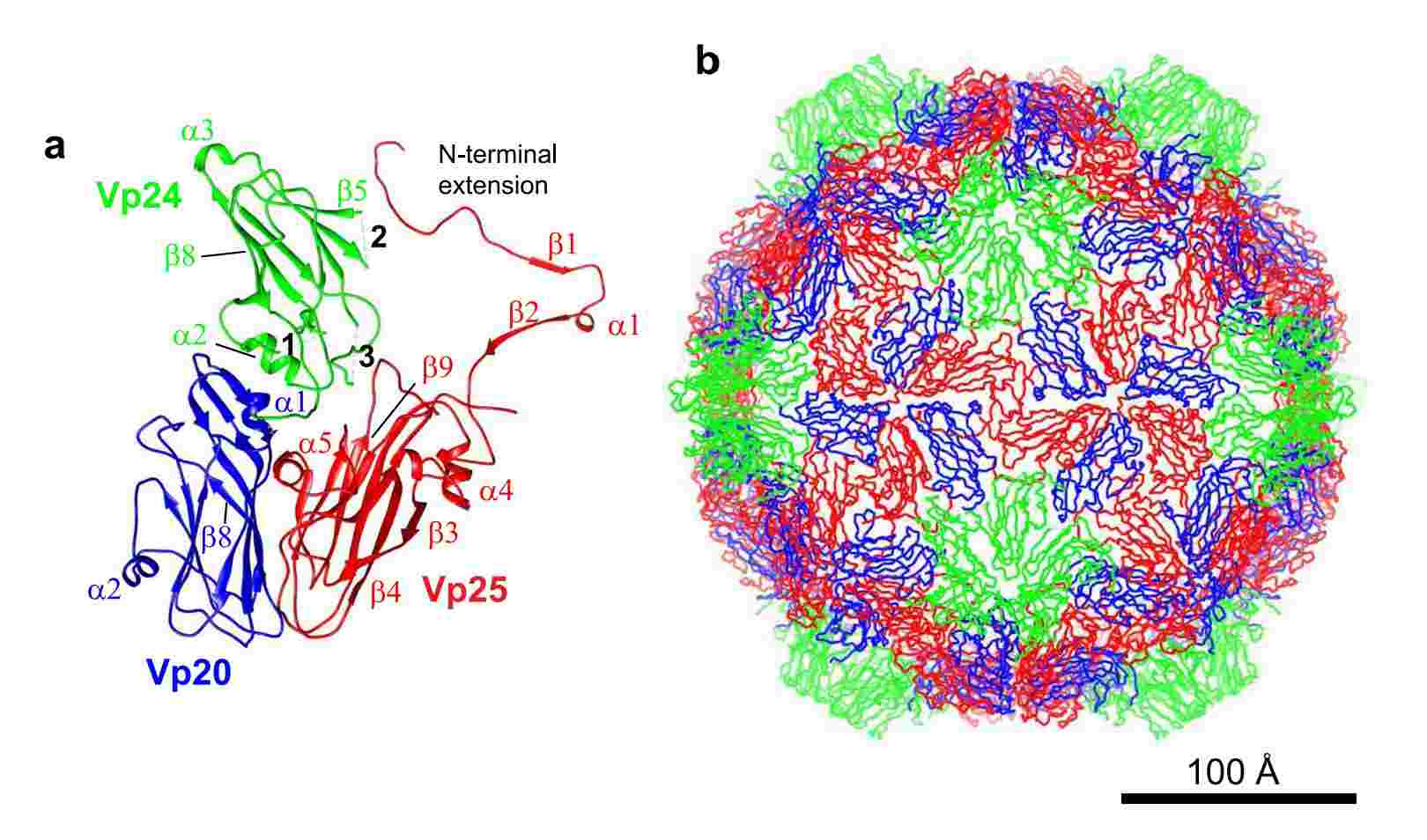 Figure 1. Structure of apple-latent spherical virus (ALSV), determined by single-particle cryo-EM. (Naitow H, et al, 2020)
Figure 1. Structure of apple-latent spherical virus (ALSV), determined by single-particle cryo-EM. (Naitow H, et al, 2020)
Structural Overview of Secoviruses
As an example, CPMV, the most widely researched member of the Secoviridae family, has empty virus-like particles (eVLP) that are currently used in biomedical and nanotechnology applications. The researchers report the crystal structure of the eVLP of CPMV, which consists of the single-stranded, positive-sense, dichotomous RNA genomes RNA-1 (6 kb) and RNA-2 (3.5 kb), as determined by X-ray crystallography at a resolution of 2.3 Å. These two RNAs are encapsulated into separate particles, and both are required for viral infection. Viral particles are approximately 310 Å and exhibit pseudo-T = 3 icosahedral symmetry. Each particle consists of 60 progenitors containing large (L) and small (S) capsid protein (CP) subunits derived from a single CP precursor (VP60) encoded by RNA-2 and processed by 24K proteinase (RNA-1-encoded). The L subunit (41 kDa) contains two jellyroll β barrels, whereas the S subunit (23 kDa) consists of a single jellyroll β barrel.
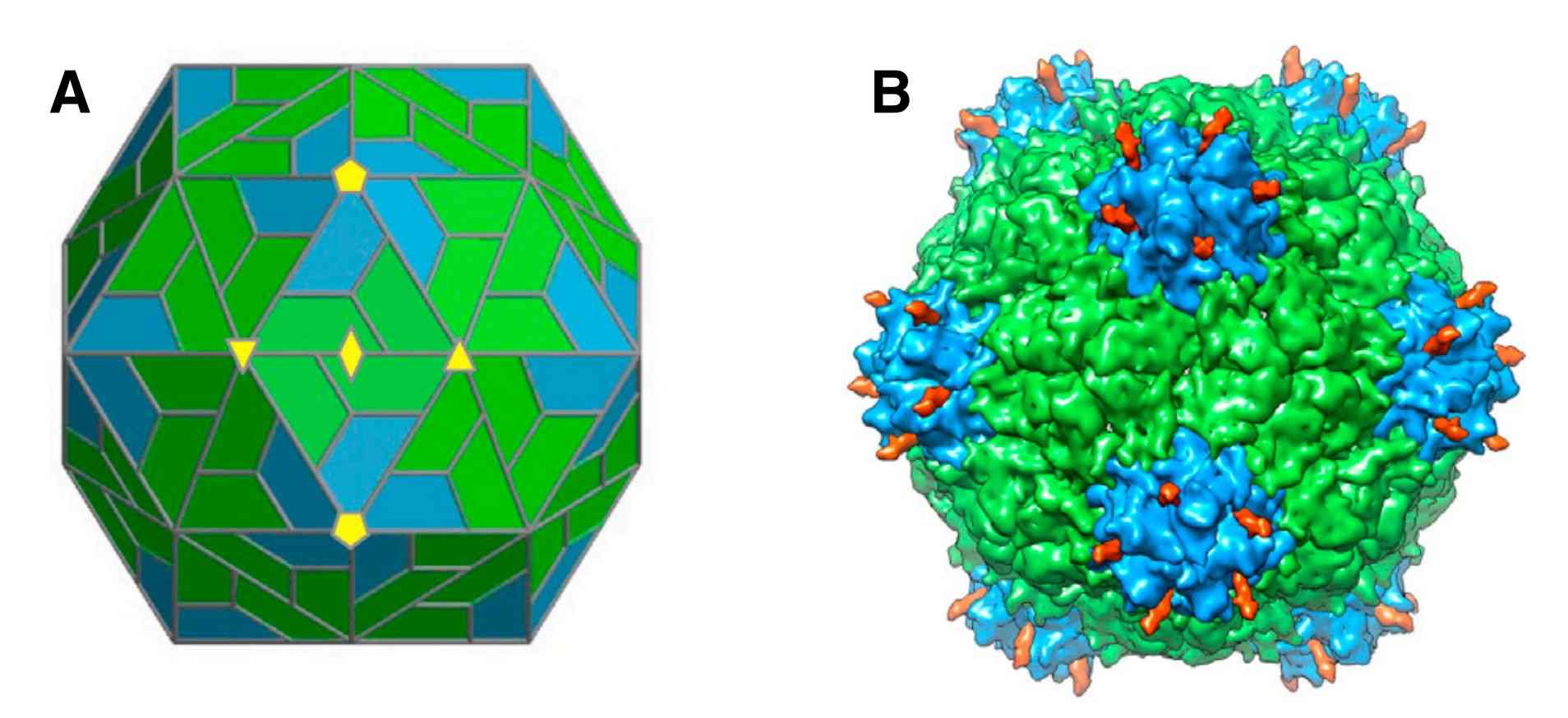 Figure 2. Structure of CPMV eVLPs. (Huynh NT, et al, 2016)
Figure 2. Structure of CPMV eVLPs. (Huynh NT, et al, 2016)
Genome Structure of Secoviruses
The genome of the virus consists mostly of two single-stranded positive-sense RNAs (RNA1 and RNA2), both essential for plant infection. P1 (encoded by RNA1) is the precursor of proteins required for replication, including deconjugating enzymes with nucleoside triphosphate-binding structural domains, proteases (Pro), and RNA-dependent RNA polymerases (Pol). One (1A) or both (X1 and X2) proteins are located upstream of the deconjugated structural domain. P2 (encoded by RNA2) includes CP, which has multiple units that form icosahedral viral particles 26-30 nm. The intercellular motor protein (MP) structural domain is immediately upstream of the CP. One (2A, required for RNA2 replication) or two (X3 and X4) proteins are located upstream of the MP. Viruses are classified according to RNA2 characteristics, including their organization and size, the phylogenetic relationship of the CP coding region, and the cleavage sites recognized by the viral protease.
Significance of Plant Virus Structure Research
Research on the structure of plant viruses is essential to plant virology, agriculture, and biotechnology.
- Understand the mechanisms of virus replication and infection.
- Resolution of virus-host interactions and pathogenesis.
- Development of antiviral strategies.
- Virus detection and diagnosis.
- Vaccine development.
- Designing viral particles to deliver antigenic or therapeutic molecules.
Creative Biostructure, as a leader in structural research of biological macromolecules, provides high-quality virus-like particles (VLPs) for virus basic research, morphogenesis, vaccine preparation, and immune characterization. The following table shows our VLPs of Secoviridae family members.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V587 | Grapevine fanleaf virus VLP (CP; coat protein Proteins) | Grapevine fanleaf virus | Plant recombinant | CP; coat protein |
| CBS-V1003 | Cowpea Mosaic Virus VLP (L; S Proteins) | Cowpea mosaic virus | Nicotiana benthamiana | L; S |
| Explore All Secoviridae Virus-like Particle Products | ||||
In addition, Creative Biostructure's viral structure elucidation services are at the forefront of cryo-electron microscopy (cryo-EM), utilizing cutting-edge platforms that set us apart in the field. Our strength lies in the superior quality and resolution of the viral structures we elucidate, allowing us to delve into the intricate details of viruses at the molecular level.
Our team of scientists has expertise in sample preparation and data analysis, ensuring that we provide clients with precise and insightful structural information. If you are interested in our services or products, please contact us. We will facilitate your progress in the fields of virology, vaccine development, and antiviral therapy.
References
- Naitow H, et al. Apple latent spherical virus structure with stable capsid frame supports quasi-stable protrusions expediting genome release. Commun Biol. 2020. 3(1): 488.
- Huynh NT, et al. Crystal Structure and Proteomics Analysis of Empty Virus-like Particles of Cowpea Mosaic Virus. Structure. 2016. 24(4): 567-575.
- Fuchs M, et al. ICTV Virus Taxonomy Profile: Secoviridae 2022. J Gen Virol. 2022. 103(12): 10.1099/jgv.0.001807.
- Hily JM, et al. Metagenomic analysis of nepoviruses: diversity, evolution and identification of a genome region in members of subgroup A that appears to be important for host range. Arch Virol. 2021. 166(10): 2789-2801.