Structural Research of Tombusviridae
Members of the Tombusviridae family are single-stranded positive-sense RNA plant viruses comprising three subfamilies. This family encompasses a wide range of soil-borne viruses, with most members infecting dicotyledonous or monocotyledonous plants, causing severe crop losses worldwide. In recent years, researchers have determined the high-resolution structures of multiple Tombusviridae family members by cryo-electron microscopy (cryo-EM), providing the molecular-level insight needed to rationally probe capsid formation and propagation, and offering a platform for developing prophylactic agrochemicals for the related essential plant virus.
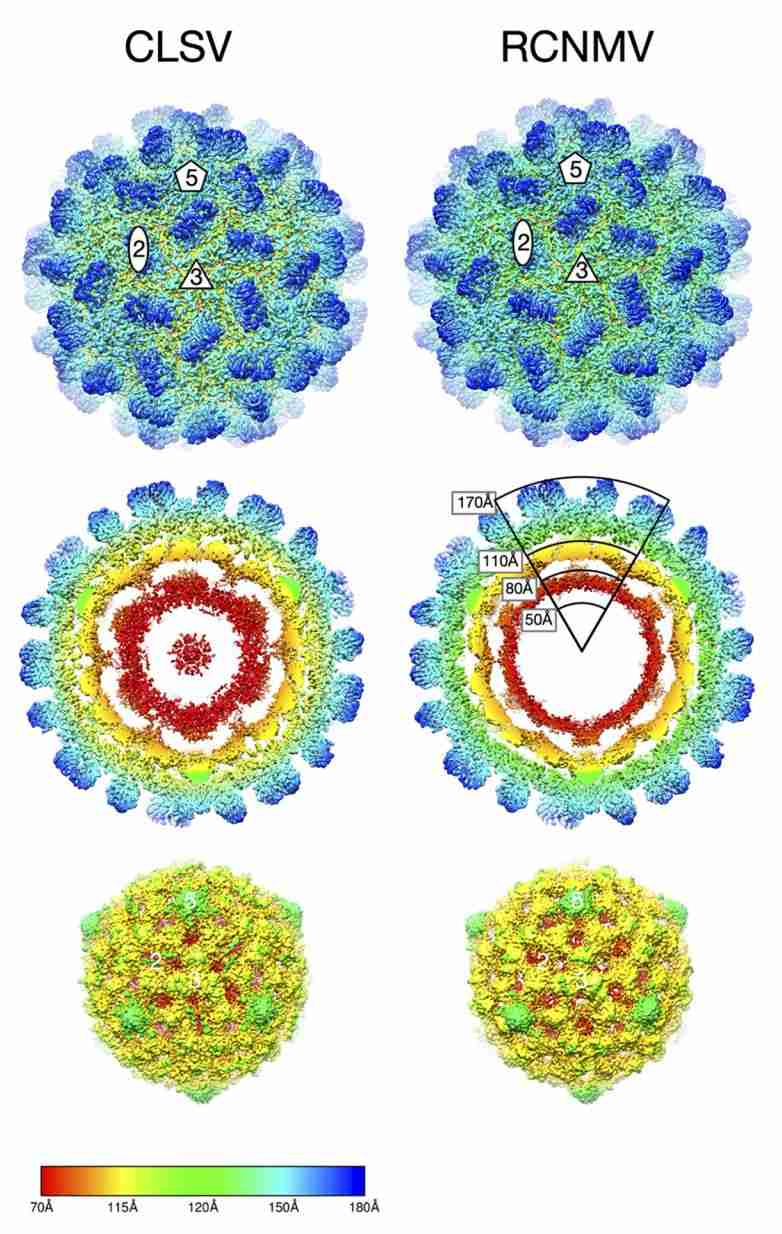 Figure 1. Cryo-EM image reconstructions of Tombusviridae virus CLSV and RCNMV. (Sherman MB, et al, 2020)
Figure 1. Cryo-EM image reconstructions of Tombusviridae virus CLSV and RCNMV. (Sherman MB, et al, 2020)
Morphological Features of Tombusviridae Virus Particles
The capsids of Tombusviridae family members exhibit T=3 icosahedral symmetry and consist of 180 identical protein subunits. Viral particles of most genera have a rounded outline and a granular surface, with a diameter of about 32-35 nm. Each subunit is folded into three distinct structural domains: the N-terminal internal structural domain R, which interacts with the RNA; the shell structural domain S, which constitutes the backbone of the capsid; and the prominent paired-aggregated C-terminal structural domain P. The S domain forms a β-barrel structure consisting of eight β-strands. The capsids of the genera Machlomovirus, Panicovirus, and Necrovirus consist of structurally similar capsid proteins and have a smooth appearance because they lack the P domain.
Progress in Structural Research on Tombusviridae
Recently, to better understand how structural details affect the mechanism of Tombusviridae virus infection, scientists used cryo-electron microscopy (cryo-EM) and image reconstruction methods to determine the structures of cucumber leaf spot virus (CLSV; genus Aureusvirus) and red clover necrotic mosaic virus (RCNMV; Dianthovirus) at 3 Å and 2 Å, respectively. The structures show that both viruses have T=3 icosahedral protein shells with a maximum diameter of ∼340 Å. Although the shell domains are homologous, the stabilizing interactions between the icosahedral triplex axis and the R domains are quite different, which may be essential for virus transmission and life cycle.
virus-like particles (VLPs) are empty viral capsids without genetic material that have structural characteristics similar to those of intact viral particles but cannot replicate and are therefore highly regarded as safe and effective candidates for vaccine development and delivery systems. Creative Biostructure offers a wide variety of VLP products to help clients explore the complex structure of viruses. This will contribute to a greater understanding of the many facets of virology and ultimately help clients design better strategies to combat pathogens.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V665 | Johnson grass chlorotic stripe mosaic virus VLP (Coat Protein Proteins) | Johnson grass chlorotic stripe mosaic virus | Plant recombinant | Coat Protein |
| Explore All Tombusviridae Virus-like Particle Products | ||||
Optical instrumentation is essential for resolving the morphology of viral particles, and Creative Biostructure has three modern techniques for visualizing viral particles, electron microscopy (EM) and X-ray crystallography. These techniques allow us to determine the high-resolution structure and gain insight into the entry mechanisms of viral particles.
Our experienced scientists and technicians can provide customized structural analysis solutions, using cutting-edge equipment and software to ensure the highest quality results and fast turnaround times. Contact us today to learn more about our structural analysis services and how we can help you advance your research in viral structural biology.
References
- Sherman MB, et al. Near-Atomic-Resolution Cryo-Electron Microscopy Structures of Cucumber Leaf Spot Virus and Red Clover Necrotic Mosaic Virus: Evolutionary Divergence at the Icosahedral Three-Fold Axes. J Virol. 2020. 94(2): e01439-19.
- Byrne MJ, et al. Combining Transient Expression and Cryo-EM to Obtain High-Resolution Structures of Luteovirid Particles. Structure. 2019. 27(12): 1761-1770.e3.