Structural Research of Mo/Wbis-MGD Oxidoreductases
Mo/W exists in a mononuclear form at the active site of enzymes that catalyze oxygen atom transfer. Many functionally identical enzymes of the DMSOR family in nature contain molybdenum cofactors (Moco)/tungsten cofactors (Wco). Enzymes containing Wco/Moco include formate dehydrogenase, nitrate reductase, formylmethane furan dehydrogenase, etc. The homologous isomers of metalloenzyme MoeA specifically participate in the insertion of Mo and W into MPT. Then, target Moco or Wco to their respective deproteinizing enzymes.
Structure and function of formate dehydrogenase(Fdh)
Most Fhds have Se cysteine residues that bind to Mo/W. Fdh-H is a monomeric enzyme, Fdh-N is a heterotrimer, and W-Fdh is a heterodimer. Fdh utilizes various electron donors/acceptors to catalyze the reversible conversion of formate to carbon dioxide. Therefore, Fdh adsorbed on the electrode can be used for electrocatalytic CO2 reduction. In addition, the complex of Fdh and hydrogenase can convert H2 and CO2 into formates for energy storage and transportation of compounds.
Structure and function of nitrate reductases
Nitrate reductase catalyzes the reduction of nitrate to nitrite. The respiratory nitrate reduction pathway involves a membrane-binding protein complex composed of three subunits, NarGHI. It participates in the oxidation-reduction of protons in biofilms. Heterotrimeric NarGHI includes catalytic subunits (NarG), subunits containing four centers (NarH), and membrane-bound heme b subunits (NarI). A pterin cofactor in the form of tricyclic pyro pterin is found in NarGH, and the side chains and corresponding rings of Ser719 exhibit different conformations.
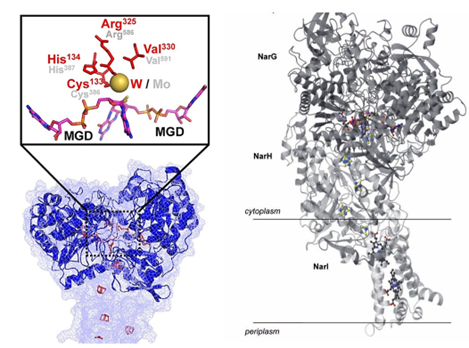 Figure 1. Fdh3A SWISS-MODEL Model (left) and overall structure of NarGHI (right). (Yang JI, et al., 2022; Moura JJ, et al., 2004)
Figure 1. Fdh3A SWISS-MODEL Model (left) and overall structure of NarGHI (right). (Yang JI, et al., 2022; Moura JJ, et al., 2004)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Virus like photosystem I | Bacillus subtilis BEST7613 | X-ray diffraction | 3.8 Å | 4L6V |
| Mitochondrial NADH: ubiquinone oxidoreductase | Yarrowia lipolytica | X-ray diffraction | 3.6 Å | 4WZ7 |
| Apo-form of NAD-dependent formate dehydrogenase in closed conformation | Moraxella sp. | X-ray diffraction | 1.96 Å | 3FN4 |
| NAD-dependent formate dehydrogenase in complex with NAD and azide | Moraxella sp. | X-ray diffraction | 1.95 Å | 2GSD |
| Holo and APO formate dehydrogenase | Pseudomonas sp. 101 | X-ray diffraction | 1.8 Å | 2NAC |
| Holo and APO formate dehydrogenase | Pseudomonas sp. 101 | X-ray diffraction | 2.05 Å | 2NAD |
| NAD+-dependent fungal formate dehydrogenase | Thermochaetoides thermophila DSM 1495 | X-ray diffraction | 1.21 Å | 6T8Z |
| Formate dehydrogenase (FDH) K47E mutant | [Candida] boidinii | X-ray diffraction | 1.7 Å | 2FSS |
| Formate dehydrogenase | Starkeya novella DSM 506 | X-ray diffraction | 2.1 Å | 7QZ1 |
| Formate dehydrogenase (FDH) C-terminal mutant | [Candida] boidinii | X-ray diffraction | 1.55 Å | 2J6I |
| NAD+-dependent fungal formate dehydrogenase | Thermochaetoides thermophila | X-ray diffraction | 1.15 Å | 6T94 |
| Apo-form of NAD-dependent formate dehydrogenase | Arabidopsis thaliana | X-ray diffraction | 1.7 Å | 3NQA |
| Formate dehydrogenase mutant V198I/C256I/P260S/E261P/S381N/S383F | Pseudomonas sp. 101 | X-ray diffraction | 2.233 Å | 6JX1 |
| Granulicella m. formate dehydrogenase (FDH) | Granulicella mallensis MP5ACTX8 | X-ray diffraction | 1.7 Å | 4XYG |
| Granulicella m. formate dehydrogenase (fdh) in complex with NADP (+) and NaN3 | Granulicella mallensis MP5ACTX8 | X-ray diffraction | 1.38 Å | 4XYB |
| Apomolybdo-NarGHI | Escherichia coli | X-ray diffraction | 2.2 Å | 1SIW |
| Nitrate Reductase A, NarGHI | Escherichia coli | X-ray diffraction | 1.9 Å | 1Q16 |
| Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | Escherichia coli | X-ray diffraction | 2 Å | 1Y4Z |
| NarGHI mutant NarI-K86A | Escherichia coli | X-ray diffraction | 1.9 Å | 1Y5I |
| NarGHI mutant NarG-H49C | Escherichia coli | X-ray diffraction | 2.3 Å | 3IR5 |
| NarGH complex | Escherichia coli | X-ray diffraction | 2 Å | 1R27 |
Table 1. Structural research of Mo/Wbis-MGD oxidoreductases.
Creative Biostructure uses X-ray crystallography to analyze the structure of Mo/Wbis-MGD oxidoreductases. We have long been committed to the study of structural biology and membrane proteins. Our experts have extensive experience in determining the structure of membrane proteins.
If you are interested in our services, please contact us and we will provide you with a professional and comprehensive solution.
References
- Yang JI, et al. A Novel NADP-Dependent Formate Dehydrogenase From the Hyperthermophilic Archaeon Thermococcus onnurineus NA1. Front Microbiol. 2022. 13:844735.
- Moura JJ, et al. Mo and W bis-MGD enzymes: nitrate reductases and formate dehydrogenases. J Biol Inorg Chem. 2004. 9(7):791-799.