In biochemistry, oxidoreductase enzymes catalyze the transfer of electrons from one molecule (the reductant, also known as the electron donor) to another (the oxidant, also known as the electron acceptor). The process typically utilizes NADP+ or NAD+ as a cofactor. Understanding the structure and function of oxidoreductases is critical to improving our ability to manipulate the oxidoreductase catalytic pathway for various applications. In recent years, significant progress has been made in structural studies of oxidoreductases, providing new insights into their catalytic mechanisms.
Structural research on membrane-bound dihydroorotate, quinone oxidoreductase (DHOQO)
Dihydroorotate, quinone oxidoreductase (DHOQO) is a flavin mononucleotide (FMN)-containing membrane-bound enzyme responsible for oxidizing dihydroorotate (DHO) to orotate with concomitant reduction of quinone to quinol. DHOQOs belong to the octet βα barrel family (βα)8, with a central core of eight parallel β chains (β1-β8) surrounded by eight α-helices (α1-α8). One side of the core has three β chains (βc-βe), often called the top side, and the other side has two (βa-βb), called the bottom side, totaling 13 β chains.
Structural studies of NADPH-cytochrome P450 oxidoreductase (CYPOR)
CYPOR is an essential flavoprotein of the microsomal cytochrome P450-monooxygenase system that transfers electrons from NADPH to FAD to FMN to members of the cytochrome P450 superfamily. It is anchored in the phospholipid bilayer by its amino-terminal membrane-bound structural domain. The structure of rat CYPOR at 2.6Å resolution confirms the presence of four separate domains, including FMN, the linker domain, FAD, and the NADPH-binding domain. The linker domain brings the two flavin structural domains close to facilitate internal electron transfer within CYPOR to interact with P450.
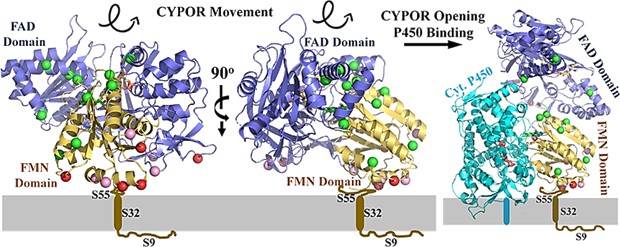 Figure 1. Orientation of CYPOR on membranes. (Xia C, et al., 2019)
Figure 1. Orientation of CYPOR on membranes. (Xia C, et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| L-amino acid deaminase | Proteus vulgaris | X-ray diffraction | 2.63 Å | 5HXW |
| A respiratory membrane-bound hydrogenase | Pyrococcus furiosus COM1 Pyrococcus furiosus DSM 3638 |
Cryo-EM single particle analysis | 3.7 Å | 6CFW |
| Particulate methane monooxygenase | Methylocystis sp. ATCC 49242 | X-ray diffraction | 3.15 Å | 4PI0 |
| Particulate methane monooxygenase | Methylocystis sp. ATCC 49242 | X-ray diffraction | 3.59 Å | 4PHZ |
| Particulate methane monooxygenase | Methylococcus capsulatus Methylococcus capsulatus str. Bath |
X-ray diffraction | 2.801 Å | 1YEW |
| Cytochrome c nitrite reductase NrfHA complex | Desulfovibrio vulgaris str. Hildenborough | X-ray diffraction | 2.3 Å | 2J7A |
| Membrane-bound respiratory [NiFe] hydrogenase in an H2-reduced condition | Hydrogenovibrio marinus | X-ray diffraction | 1.18 Å | 3AYX |
| Membrane-bound respiratory [NiFe] hydrogenase in an air-oxidized condition | Hydrogenovibrio marinus | X-ray diffraction | 1.22 Å | 3AYZ |
| Membrane-bound respiratory [NiFe]-hydrogenase in a ferricyanide-oxidized condition | Hydrogenovibrio marinus | X-ray diffraction | 1.32 Å | 5Y34 |
| Membrane-bound alcohol dehydrogenase | Gluconobacter oxydans Gluconobacter oxydans 621H |
Cryo-EM single particle analysis | 2.5 Å | 8GY2 |
| Krypton derivatization of an O2-tolerant membrane-bound [NiFe] hydrogenase | Cupriavidus necator | X-ray diffraction | 1.47 Å | 5D51 |
| Membrane-bound manganese superoxide dismutase | Nostoc sp. PCC 7120 = FACHB-418 | X-ray diffraction | 2 Å | 1GV3 |
| Hydrogenase-1 in complex with cytochrome b | Escherichia coli K-12 | X-ray diffraction | 3.3 Å | 4GD3 |
| Gamma-alpha subunit complex from glucose dehydrogenase | Burkholderia cepacia | X-ray diffraction | 2.6 Å | 6A2U |
| H2-reduced structure of hydrogenase-1 | Escherichia coli | X-ray diffraction | 1.47 Å | 3UQY |
| A novel membrane-bound 6-phosphogluconate dehydrogenase | Gluconacetobacter diazotrophicus PA1 5 | X-ray diffraction | 1.87 Å | 6VPB |
| Membrane-bound aldehyde dehydrogenase | Gluconobacter oxydans | Cryo-EM single particle analysis | 2.7 Å | 8GY3 |
| O2-tolerant MBH-P242C in its reduced state | Cupriavidus necator H16 | X-ray diffraction | 1.62 Å | 7ODG |
| Cytochrome bd-II type oxidase with bound aurachin D | Escherichia coli BW25113 | Cryo-EM single particle analysis | 3 Å | 7OSE |
| MdbA protein, a thiol-disulfide oxidoreductase | Corynebacterium diphtheriae NCTC 13129 | X-ray diffraction | 1.77 Å | 5C00 |
| The lipoyl domain of the chimeric dihydrolipoyl dehydrogenase P64K | Neisseria meningitidis | SOLUTION NMR | / | 1GJX |
| R. rubrum transhydrogenase domain III bound to NADPH | Rhodospirillum rubrum | X-ray diffraction | 2.4 Å | 1PNQ |
| Oxidized active mutant of (S)-mandelate dehydrogenase | Pseudomonas putida Spinacia oleracea |
Cryo-EM single particle analysis | 1.35 Å | 1P4C |
Table 1. Structural research of oxidoreductases.
Creative Biostructure is dedicated to protein structure research. Our experts have extensive experience in the structural determination of membrane proteins. We provide NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography platforms to probe protein structures. We help our customers to explore the potential functions of oxidoreductases.
In addition to membrane protein structure determination, we can accurately analyze other biomolecules including, but not limited to, ribosomes, nucleic acids, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us for a professional solution strategy.
References
- Xia C, et al. Structural and Functional Studies of the Membrane-Binding Domain of NADPH-Cytochrome P450 Oxidoreductase. Biochemistry. 2019. 58(19):2408-2418.
- Sousa FM, et al. Investigating the amino acid sequences of membrane bound dihydroorotate: quinone oxidoreductases (DHOQOs): Structural and functional implications. Biochim Biophys Acta Bioenerg. 2021.1862(1):148321.
- Jensen K, Møller BL. Plant NADPH-cytochrome P450 oxidoreductases. Phytochemistry. 2010. 71(2-3):132-141.
