DNA Structure
Deoxyribonucleic acid (abbreviated as DNA) is a type of nucleic acid that carries genetic information in many organisms. It is essential for protein production, cell regulation, metabolism, and reproduction. Large compressed DNA molecules with related proteins (called chromatin) are mainly present in the nucleus. Several cytoplasmic organelles such as mitochondria also contain DNA molecules. "DNA Structure" can mean something as simple as the sequence of deoxyribonucleotides in a DNA fragment, or something as complicated as the way of folding a DNA molecule and how it interacts with other molecules.
Deoxyribonucleotide
As mentioned above, DNA molecules are composed of deoxyribonucleotides, each of which consists of three parts: a pentose sugar called deoxyribose, a phosphate group, and a nitrogenous base. DNA utilizes four types of nitrogenous bases, including adenine (A), guanine (G) cytosine (C), and thymine (T). Therefore, the DNA consists of four kinds of deoxyribonucleotides, respectively, deoxyadenosine monophosphate (dAMP), deoxyguanosine monophosphate (dGMP), deoxycytidine monophosphate (dCMP), and deoxythymidine monophosphate (dTMP). The dinucleotide of DNA is formed by covalently linking the 5'-phosphate group of one deoxynucleotide to the 3'-hydroxyl group of another deoxynucleotide to form a phosphodiester bond. The primary structure of a DNA molecule refers to the sequence of its deoxyribonucleotides (i.e., base sequence), and the way these are covalently bonded to each other.
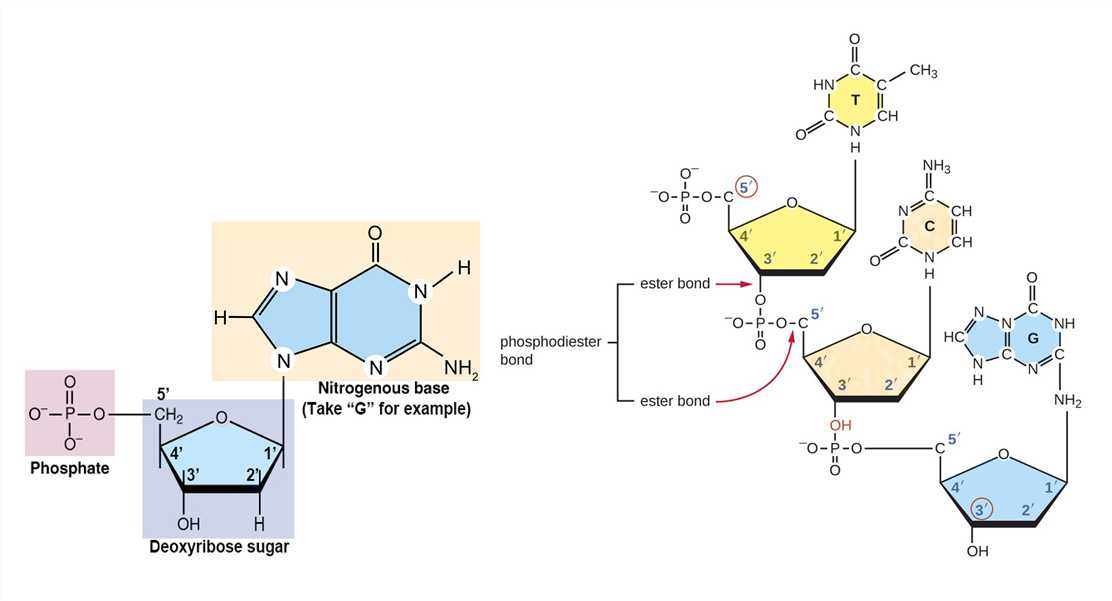 Figure 1. The composition of deoxyribonucleotide and phosphodiester bond.
Figure 1. The composition of deoxyribonucleotide and phosphodiester bond.
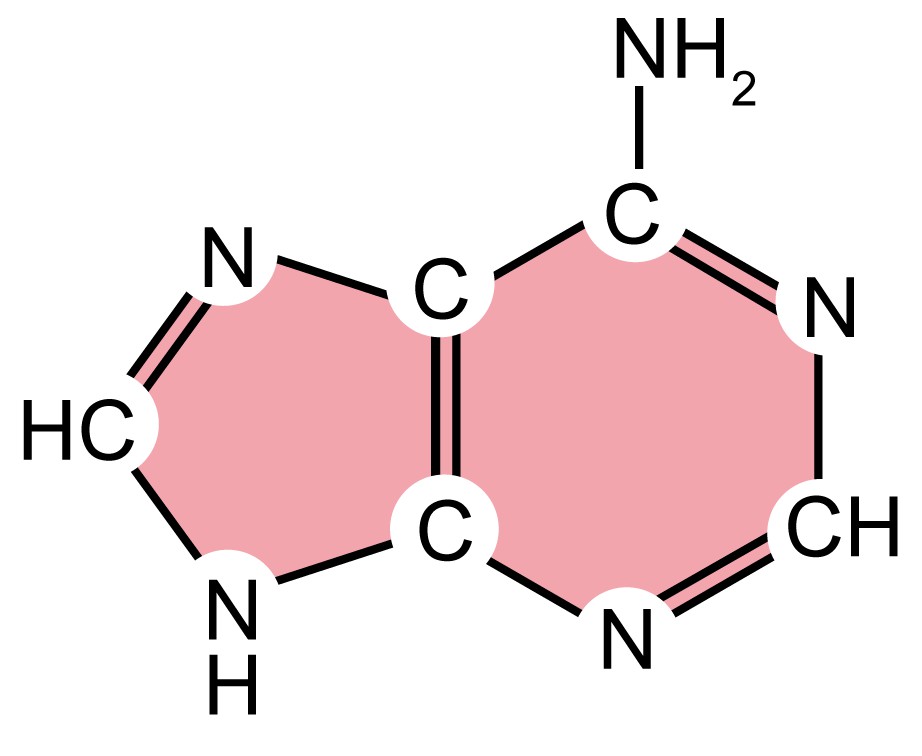
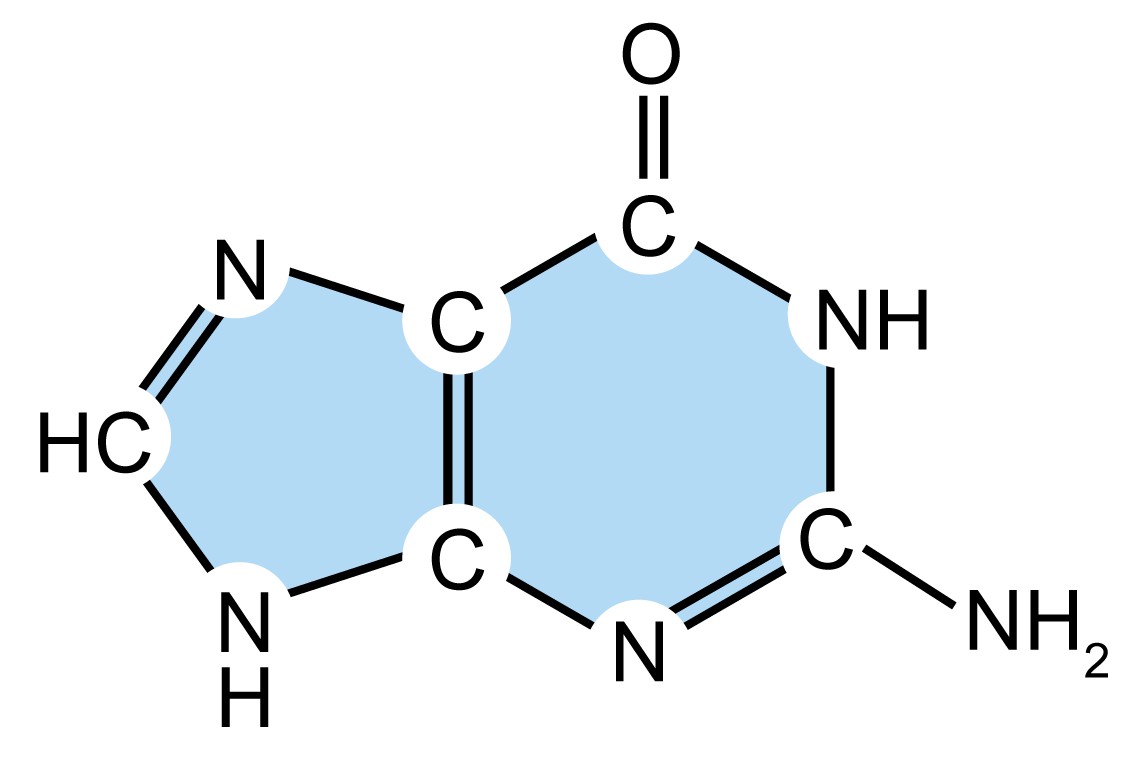
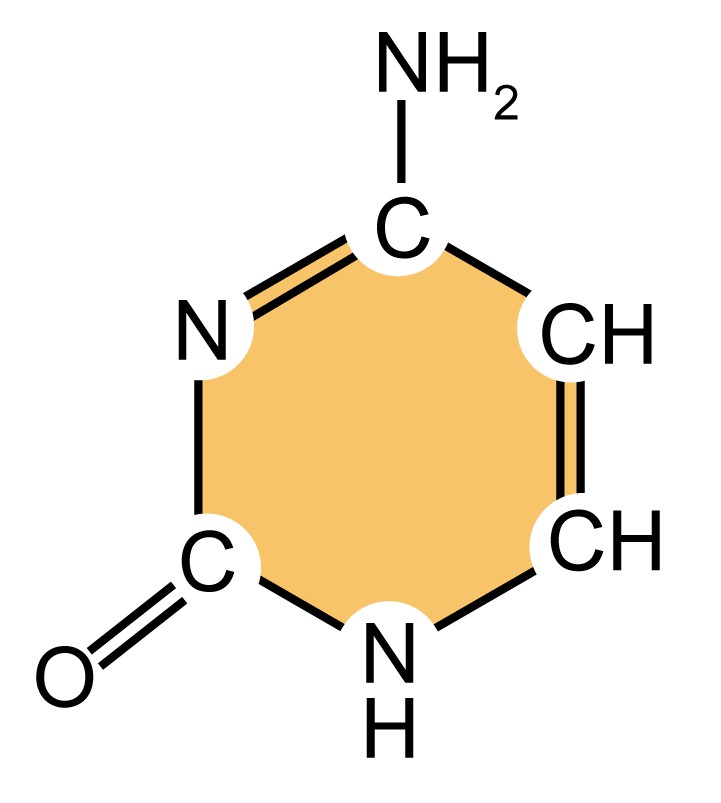
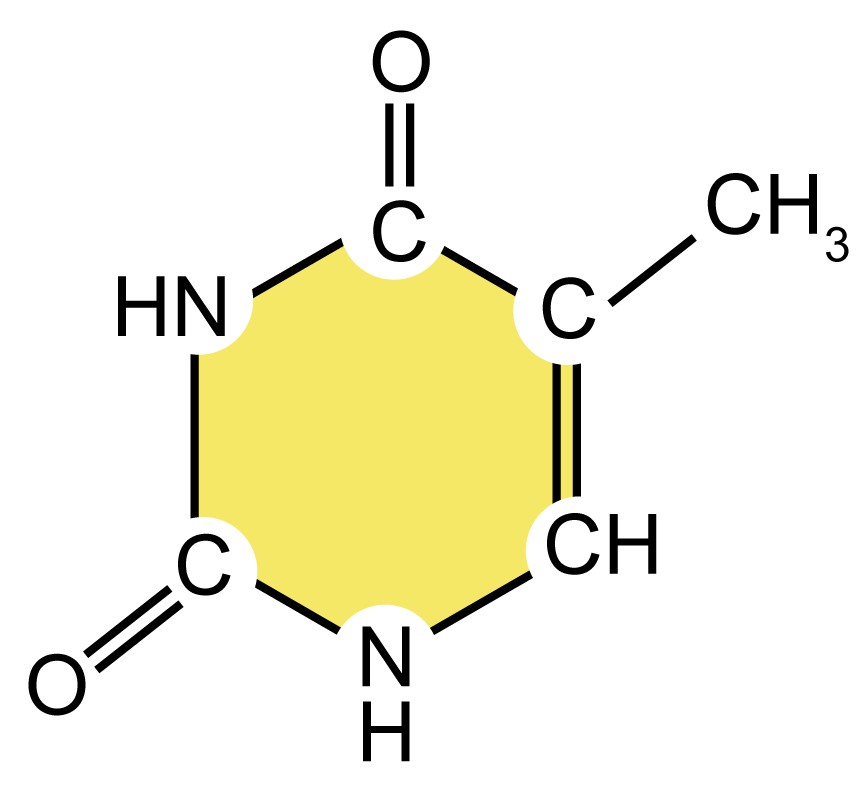
| Name (abbreviation) | Chemical structure | Classification | Characteristics |
| adenine (A) |
| Purine
| They have a double-ring structure with a six-carbon ring fused to a five-carbon ring. |
| guanine (G) |
| ||
| cytosine (C) |
| Pyrimidine
| They are smaller nitrogenous bases that have only a six-carbon ring structure. |
| thymine (T) |
|
Table 1. Four types of nitrogenous bases in DNA.
- Chargaff's rule
In the 1950s, biochemist Erwin Chargaff discovered that although the four kinds of deoxyribonucleotides (containing four different nitrogenous bases) were not equal in content, the amount of dAMP is always equal to the amount of dTMP, and the amount of dCMP is always equal to dGMP. This finding is termed Chargaff's rule and turned out to be crucial for revealing the DNA double helix model.
The DNA Double Helix
DNA can exist as a single strand or a double helix. In 1953, Watson and Crick succeeded in inferring the double helix structure of DNA using X-ray crystallography, and then the principle of complementary base-pairing was also proposed. The DNA double helix is composed of a pair of polynucleotide strands coiled around a common central axis. All four nitrogenous bases are confined within the double helix, held in place by hydrogen bonds connecting the complementary bases on the two strands. The phosphate of one deoxynucleotide is covalently connected to the deoxyribose of the next deoxynucleotide. The deoxyribose-phosphate backbone of DNA is outside the double helix. The hydrogen bonds between phosphates lead to DNA strand distortion. Although the DNA double helix exists in many possible conformations, including A-DNA, B-DNA, and Z-DNA forms, only B-DNA and Z-DNA have been directly observed in functional organisms. The main form of DNA double helix, B-DNA, has a broad major groove and a narrow minor groove extending around the helix along the entire length of the molecule.
The orientation of the polynucleotide can be defined by the phosphodiester bond between two linked deoxynucleotides. In the deoxyribose and phosphate moieties, the phosphate group is attached to the 5' carbon of one deoxyribose and the hydroxyl group of the 3' carbon in the next deoxyribose, thus successfully linking the deoxynucleotides. One strand has different ends: the end of the 5' carbon not connected to another deoxynucleotide is referred to as the 5' end, and the other end is called the 3' end. The two DNA strands in the DNA double helix are antiparallel to each other.
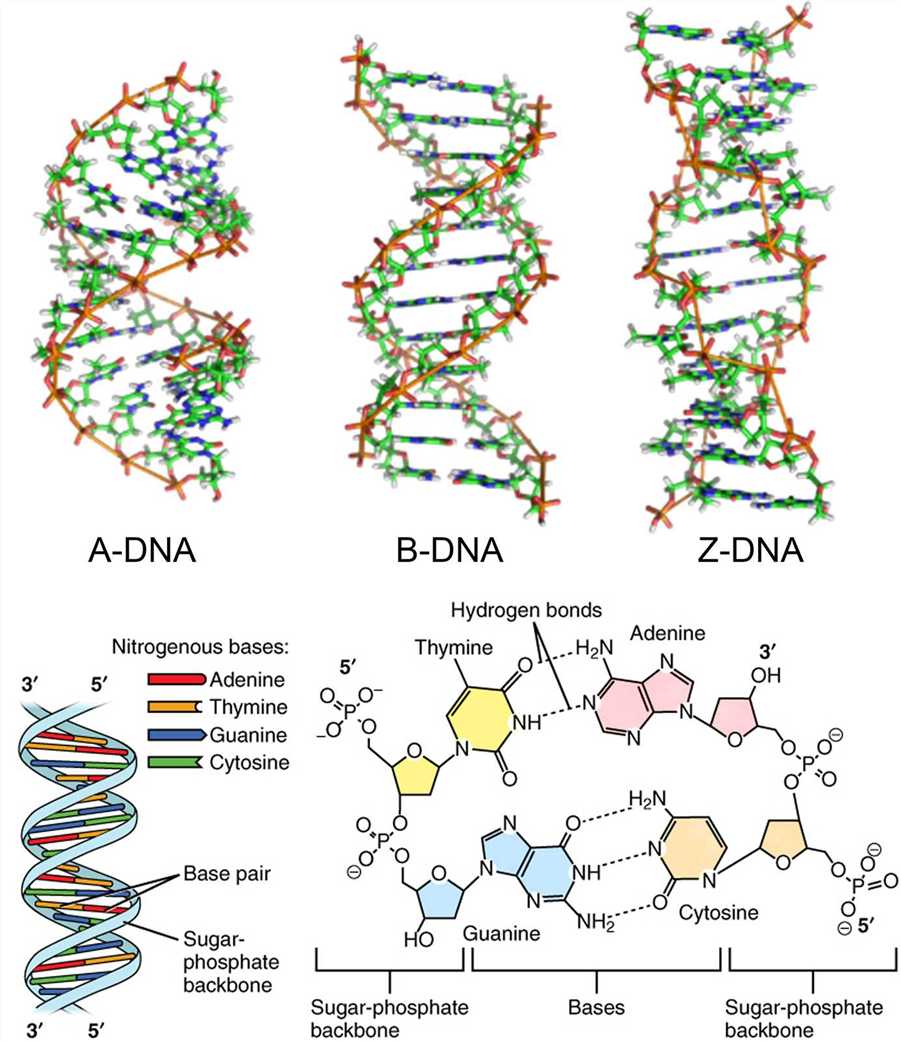 Figure 2. The DNA double helix.
Figure 2. The DNA double helix.
- Complementary base-pairing
Each of the four nitrogenous bases of DNA contains a unique form of hydrogen bond receptor and donor, thus only certain base pairs are allowed. In double-stranded DNA, the adenine-thymidine (A-T) base pair contains two hydrogen bonds, while the guanine-cytosine (G-C) base pair contains three hydrogen bonds, and the hydrophobic binding between the nitrogenous bases provides stability of the DNA molecule. These interactions are known as Watson-Crick base pairs. The ability of nucleic acid molecules to form base pairs is the basis of all life, and one nucleic acid strand must be bound to another strand by complementary base pairing. Additionally, a single strand can be utilized as a template for synthesizing another complementary strand.
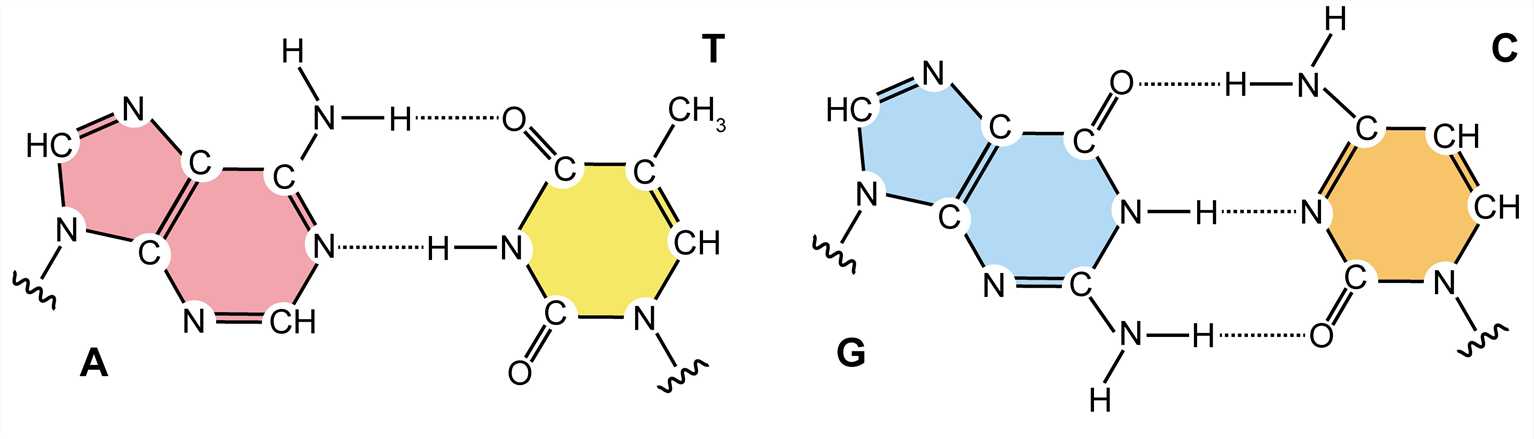 Figure 3. Watson-Crick base pairs.
Figure 3. Watson-Crick base pairs.
DNA Nanostructure
In 1982, Seeman proposed that DNA can form specific structures through complementary base-pairing, and individual structures can form complicated two-dimensional or three-dimensional structures through sticky ends. This indicates that DNA is no longer just genetic material, but also can be folded into specific two-dimensional and three-dimensional structures to construct various functional structures and nanodevices as a natural nanomaterial. Lots of DNA nanostructures have been produced and a wide range of potential applications have been proposed, such as DNA cages for drug delivery, protein crystal templates, medical diagnosis, and DNA computing systems.
- Natural DNA nanostructure: G-quadruplex structure
G-quadruplex (G4) is different from the traditional DNA double helix formed by the principle of complementary base pairing. It is a stacked nucleic acid structure that can form within specific repetitive G‑rich DNA sequences. In 1962, Gellert and colleagues demonstrated that guanylic acids can assemble into tetrameric structures. In these tetramers, four guanine molecules form a square planar arrangement (G-quartet) in which each guanine is bonded to the two adjacent guanines by hydrogen bonds. G-rich DNA can form a G-quartet under the stabilization of certain low-valent cations, and because of the continuous distribution of G bases in the DNA strand, the G-quartets can be stacked to form a G4 structure. The G4 structure is mainly distributed in the guanine-rich region of telomere DNA and promoters of various biological cells, and it is a natural DNA nanostructure.
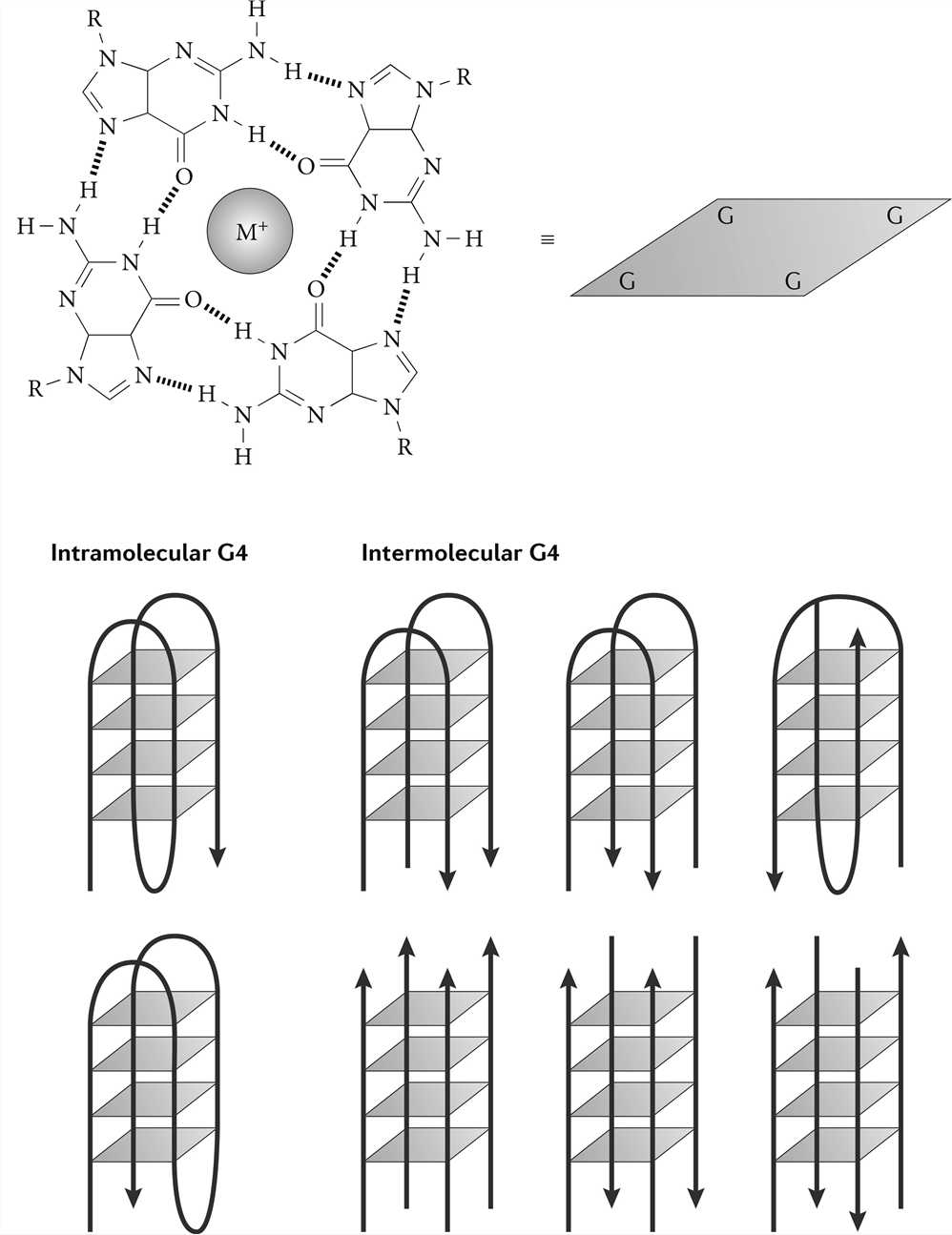 Figure 4. G-quadruplex structure.
Figure 4. G-quadruplex structure.
- Artificial DNA nanostructure: DNA origami
DNA origami utilizes the special structure of DNA molecules and the principle of complementary base-pairing to fold a specific region of a long circular single-stranded DNA and fix it with a short chain to construct the desired structure. It is done in five main steps: a) construct the desired geometric model and scaffold strands; b) fold the special area of the scaffold strands; c) combine the excess staple strands with the scaffold strands; d) adjust and merge the staples; e) annealing results in the expected DNA nanostructure. Complex two-dimensional structures and three-dimensional structures have been obtained using DNA origami.
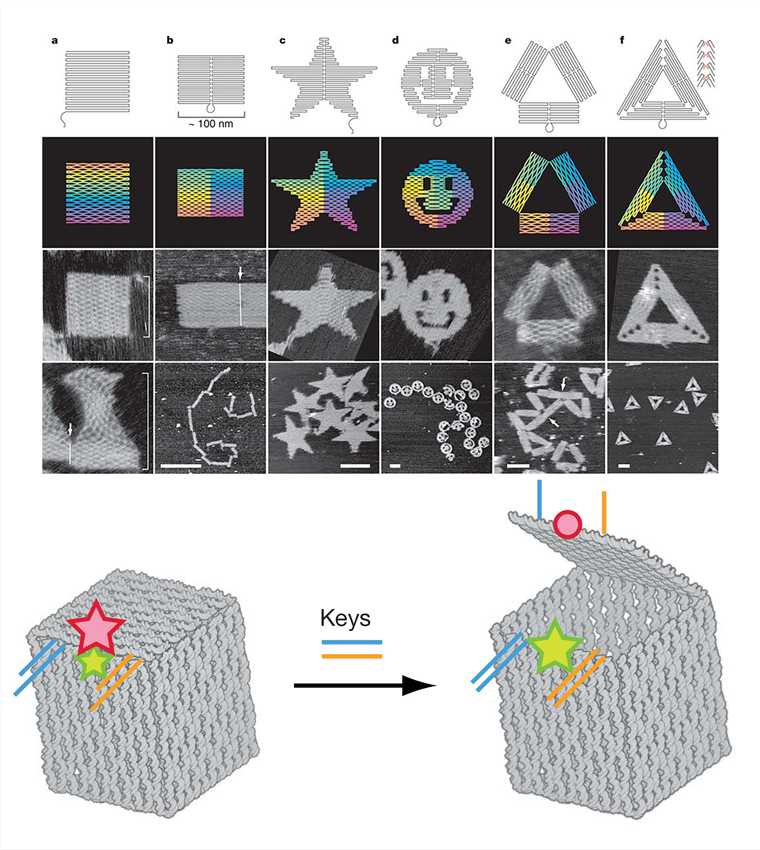 Figure 5. DNA origami shapes and nanoscale DNA box.
Figure 5. DNA origami shapes and nanoscale DNA box.
- Functional DNA nanostructure: Aptamer
The aptamer is a functional DNA nanostructure that has been artificially synthesized and screened. It is a type of DNA or RNA sequence screened by the systematic evolution of ligands by exponential enrichment technology (SELEX). The sequence can form special three-dimensional structures (e.g., hairpins, fake knots, and convex rings) through intermolecular forces such as hydrogen bonding, van der Waals force, and hydrophobic interaction, to specifically and efficiently combine with various targets (such as metal ions, small molecules, proteins, and cells). Because the aptamer can bind to other molecules specifically and efficiently, it is similar to the effect of traditional antibodies to a certain extent. Therefore, it is also called "chemical antibody".
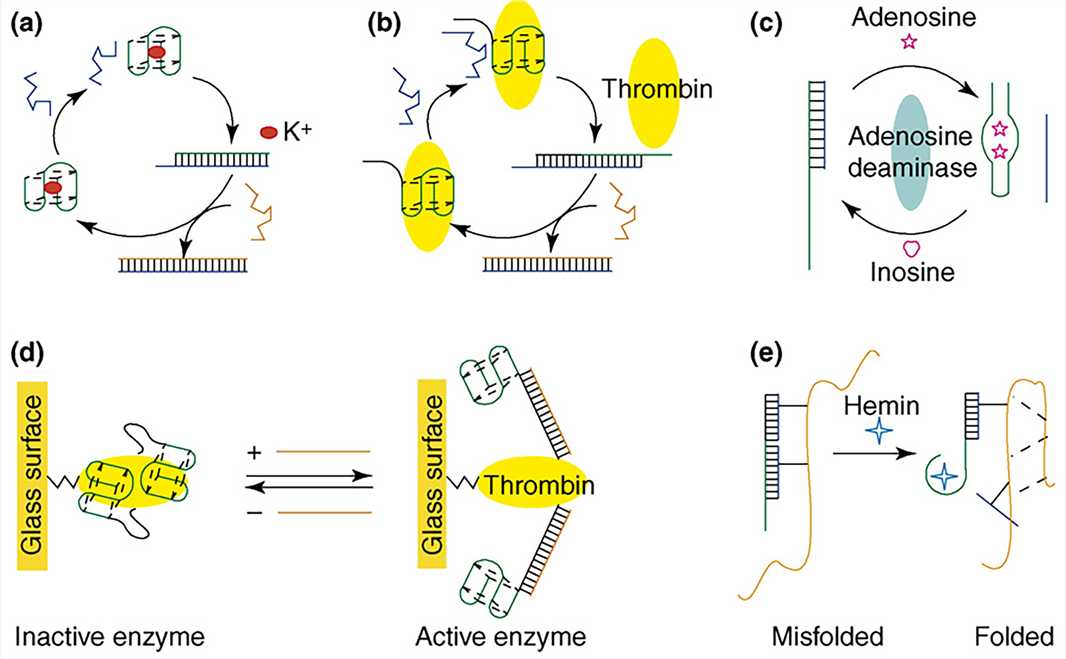 Figure 6. Aptamer-based DNA devices.
Figure 6. Aptamer-based DNA devices.
For practical applications of DNA nanostructures, it is necessary to verify that the three-dimensional structure is precisely produced according to the design, which requires high resolution, at least enough to resolve the DNA helix. However, given its extremely small size, the inspection and validation of its precise three-dimensional constructions are particularly challenging. Remarkably, Cryo-EM has been shown to be exploited to obtain DNA nanostructure information of sufficient resolution.
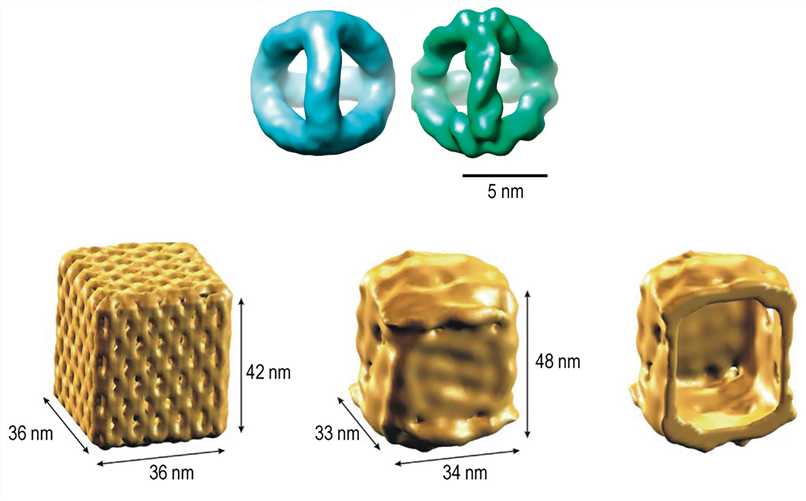 Figure 7. 3D density maps of the DNA tetrahedron and nanoscale DNA box revealed by Cryo-EM reconstruction.
Figure 7. 3D density maps of the DNA tetrahedron and nanoscale DNA box revealed by Cryo-EM reconstruction.
Creative Biostructure is specialized in the field of structural biology, and we provide contract services for the structural analysis of DNA samples.
References
- Bochman M L, et al. DNA secondary structures: stability and function of G-quadruplex structures. Nature Reviews Genetics. 2012. 13(11): 770-780.
- Rothemund P W K. Folding DNA to create nanoscale shapes and patterns. Nature. 2006. 440(7082): 297-302.
- Andersen E S, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009. 459(7243): 73-76.
- Lu Y, Liu J. Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers. Current Opinion in Biotechnology. 2006. 17(6): 580-588.
- Kato T, et al. High-resolution structural analysis of a DNA nanostructure by cryoEM. Nano Letters. 2009. 9(7): 2747-2750.