Structural Research of Urea Transporters
Urea is a critical molecule in a variety of physiological processes, including the urea cycle and regulation of urine output, and urea transporters (UTs) are a class of integral membrane proteins that mediate the passive diffusion of urea along concentration gradients. In mammals, UTs are expressed in a variety of tissues, including the kidney, liver, and brain. Its function is best understood in the kidney, where it helps to maintain the high interstitial urea concentration required to limit the rate of water loss.
Primary structure of UT
The amino acid sequences of members of the UT family share many common features. UT typically contains 10 predicted transmembrane helices or, in the case of the UT-A1 isoform, 20 predicted transmembrane helices from the two UT structural domains in tandem. Within each UT structural domain, regions corresponding to the first and last five transmembrane helices show significant homology, suggesting that the protein may have arisen from the replication of a five-transmembrane-helix protein. A comparison of the UT sequences revealed that each of the five transmembrane repeats contains the LPXXTXPF conserved signature motif, which is thought to play an essential role in urea permeation.
Structural research on human UT-A and UT-B
UT-A and UT-B mediate the reabsorption process of urea excretion in humans and are targets for the development of novel salt-preserving diuretics. In recent years, researchers have used graphene lattices to overcome the inherent problems of selective orientation and low yields to determine the high-resolution structures of human UT-A and UT-B by protein crystallography and cryo-electron microscopy (cryo-EM). The structures show that UT-A and UT-B are in a trimeric arrangement with 10 TM helices and two pore (P) helices per subunit. The channel-like pore for urea transport is open and consists of V69, T177, L292, and F342. Compared to other UTs, UT-A showed more negative surface charge on its extracellular side, and the extracellular surface of UT-B is only slightly negatively charged.
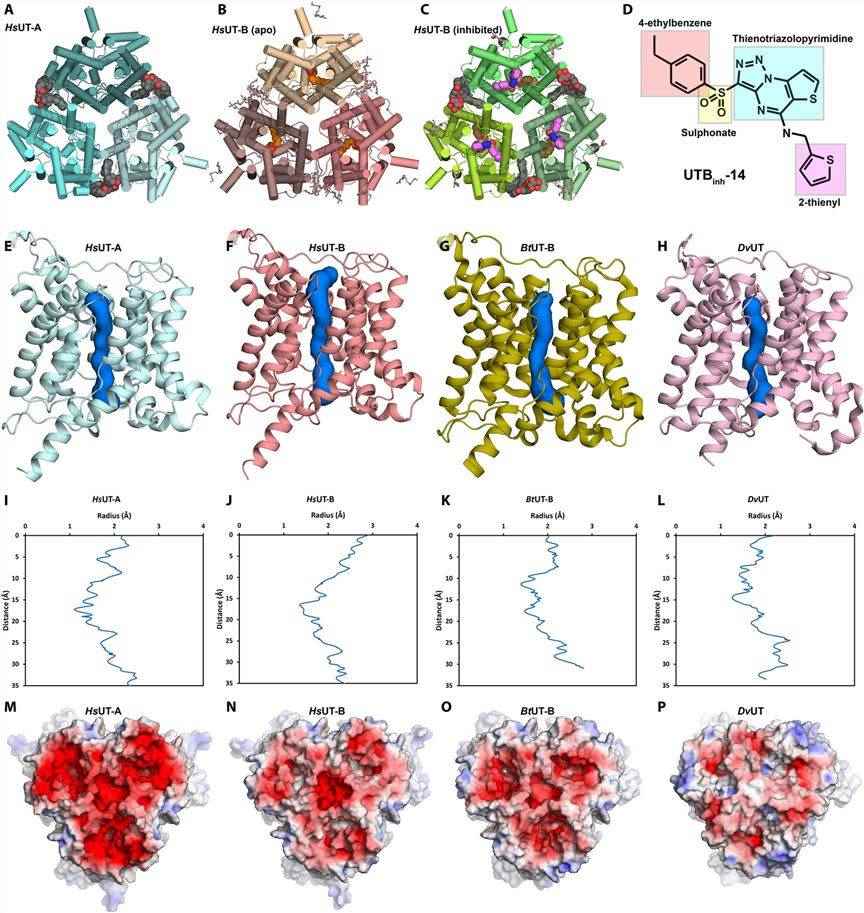 Figure 1. The general structure of HsUT-A and HsUT-B. (Chi G, et al., 2023)
Figure 1. The general structure of HsUT-A and HsUT-B. (Chi G, et al., 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Urea transporter | Nitratidesulfovibrio vulgaris str. Hildenborough | X-ray diffraction | 2.3 Å | 3K3F |
| UT-B urea transporter | Bos taurus | X-ray diffraction | 2.36 Å | 4EZC |
| UT-B urea transporter bound to selenourea | Bos taurus | X-ray diffraction | 2.5 Å | 4EZD |
| Urea transporter bound to 1,3-dimethylurea | Nitratidesulfovibrio vulgaris str. Hildenborough | X-ray diffraction | 2.4 Å | 3K3G |
| Urea channel SLC14A1/UT1 | Homo sapiens | X-ray diffraction | 2.398 Å | 6QD5 |
| Urea transporter in the P3(1) space group | Nitratidesulfovibrio vulgaris str. Hildenborough | X-ray diffraction | 3.862 Å | 3ME1 |
| F80A mutant of the Urea Transporter | Nitratidesulfovibrio vulgaris str. Hildenborough | X-ray diffraction | 2.65 Å | 3M6E |
| Urea transporter UT-B/UT1 in complex with inhibitor UTBinh-14 | Homo sapiens | Cryo-EM single particle analysis | 2.6 Å | 8BLP |
| Urea channel in closed state | Helicobacter pylori J99 | Cryo-EM single particle analysis | 2.7 Å | 6NSJ |
| Urea channel in open state | Helicobacter pylori J99 | Cryo-EM single particle analysis | 2.7 Å | 6NSK |
| Urea transporter UT-A (N-terminal domain model) | Homo sapiens | Cryo-EM single particle analysis | 2.9 Å | 8BLO |
| UrtA in complex with urea and calcium | Prochlorococcus marinus str. MIT 9313 | X-ray diffraction | 1.6 Å | 8HIC |
| UrtA in complex with urea and calcium | Synechococcus sp. CC9311 | X-ray diffraction | 1.973 Å | 7S6E |
| UrtA1 in complex with urea and calcium | Parasynechococcus marenigrum WH 8102 | X-ray diffraction | 1.8 Å | 7S6F |
Table 1. Structural research of the urea transporters.
Creative Biostructure is a preeminent company providing innovative protein structure analysis services. We make full use of cutting-edge structural biology techniques to research the structure and function of membrane proteins. Our team of scientists has extensive experience in X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy, which gives us the ability to provide detailed membrane protein structure analysis services.
Our membrane protein structure analysis services provide invaluable assistance to researchers in unraveling the complex molecular mechanisms of urea transporters. With our unrivaled expertise in the field of structural biology, we can provide an accurate and detailed understanding of the structure and function of membrane proteins. If you would like to learn more about our services, please feel free to contact us. Our team of scientists is ready to provide the best solution for your project.
References
- Chi G, et al. Structural characterization of human urea transporters UT-A and UT-B and their inhibition. Sci Adv. 2023. 9(39): eadg8229.
- Levin EJ, Zhou M. Structure of urea transporters. Subcell Biochem. 2014. 73: 65-78.