Structural Research of Polysaccharide Utilization Proteins
Polysaccharide utilization proteins (PUPs) are enzymes encoded by polysaccharide utilization sites (PULs) that sense nutrients and facilitate glycan digestion. Members of the human gut microbiome, especially Anabaena, contain high levels of PUPs that aid in the digestion and absorption of carbohydrates in the body. Research on the structure of PUPs can provide insights into fundamental biological processes, offering potential applications in biotechnology and human health. It has broad implications for various fields ranging from basic science to pharmaceutical research.
PUP crystallization and model construction process
Protein crystals are grown by mixing the protein solution with the reservoir solution using the sitting-drop vapor diffusion method. The crystals are flash-cooled by immersion in liquid nitrogen. Data is collected by X-ray diffraction and is treated using XDS. Then, data from SeMet-labelled protein crystals are subjected to phase determination and automated model construction using PHENIX's AutoSol process. Finally, manual model reconstruction and crystallographic refinement are performed by Coot and Refmac5.
Structural analysis of β-L-arabinobiosidase (HypBA2)
β-L-arabinobiosidase (HypBA2) belongs to the glycoside hydrolase family 121 (GH121) and is a type of PUP. Using X-ray diffraction to demonstrate the crystal structure of the catalytic region of HypBA2 at a resolution of 1.85 Å, the researchers found that the HypBA2 consists of a central catalytic (α/α) 6-barrel domain and two flanking (N-terminal and C-terminal) β-sandwich domains. The pocket in the catalytic domain appears to be suitable for accommodating the β-Ara 2 disaccharide. The three acidic residues Glu383, Asp515, and Glu713 located in this pocket are fully conserved among all members of GH121.
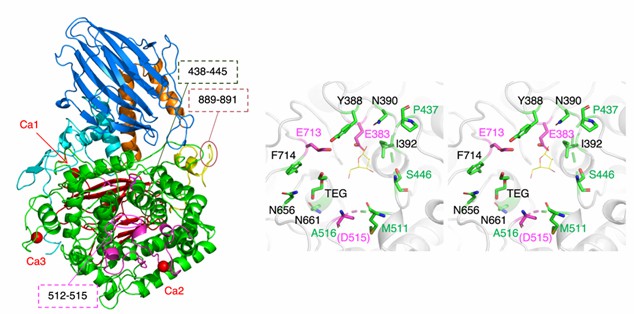 Figure 1. The overall structure of CΔ1049 (left) and active site view (right) of HypBA2. (Saito K, et al., 2020)
Figure 1. The overall structure of CΔ1049 (left) and active site view (right) of HypBA2. (Saito K, et al., 2020)
| Protei | Organism | Method | Resolutio | PDB Entry ID |
| Beta-galactosidase | Escherichia coli K-12 | Cryo-EM single particle analysis | 3.3 Å | 8BK7 |
| Beta-galactosidase | Escherichia coli K-12 | Cryo-EM single particle analysis | 2.9 Å | 8BK8 |
| Beta-galactosidase | Escherichia coli | X-ray diffraction | 2.5 Å | 4V40 |
| Protein in the Glycosyl Hydrolase Family 38 | Enterococcus faecalis V583 | X-ray diffraction | 2.55 Å | 3LVT |
| Beta-xylosidase | Thermotoga maritima MSB8 | X-ray diffraction | 1.89 Å | 7ZEQ |
| Alpha-mannosidase | Enterococcus faecalis V583 | X-ray diffraction | 2.4 Å | 5KBP |
| Beta-xylosidase in complex with methyl-beta-D-xylopyranoside | Thermotoga maritima MSB8 | X-ray diffraction | 1.46 Å | 7ZDY |
| Glycosil hydrolase family 3 N-terminal domain protein | Bacteroides intestinalis DSM 17393 | X-ray diffraction | 2.2021 Å | 5TF0 |
| Glycoside Hydrolase BglX | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 1.8 Å | 6R5I |
| PL6 family chondroitinase B | Pseudopedobacter saltans DSM 12145 | X-ray diffraction | 1.58 Å | 7O78 |
| PL6 family polysaccharide lyase Pedsa3628 | Pseudopedobacter saltans DSM 12145 | X-ray diffraction | 1.93 Å | 7O79 |
| PL6 alginate lyase BcAlyPL6 | Bacteroides clarus | X-ray diffraction | 2.213 Å | 7DMK |
| PL6 family alginate lyase AlyGC | Paraglaciecola chathamensis | X-ray diffraction | 2.194 Å | 5GKD |
Table 1. Structural research of polysaccharide utilization proteins.
As a pioneer in the field of structural analysis services, Creative Biostructure has an outstanding team of experts who specialize in the in-depth structural and functional analysis of polysaccharide utilization proteins using cutting-edge techniques such as cryo-electron microscopy (cryo-EM), X-ray crystallography, and NMR spectroscopy. From protein expression, purification to structure determination, we provide clients with one-stop comprehensive services. If you are interested in our services, please feel free to contact us for more details. We are firmly committed to providing high-quality, accurate, and timely structural analysis results.
References
- Saito K, et al. Crystal structure of β-L-arabinobiosidase belonging to glycoside hydrolase family 121. PLoS One. 2020.15(6):e0231513.
- Kappelmann L, et al. Polysaccharide utilization loci of North Sea Flavobacteriia as basis for using SusC/D-protein expression for predicting major phytoplankton glycans. ISME J. 2019.13(1):76-91.
- Feng J, et al. Polysaccharide utilization loci in Bacteroides determine population fitness and community-level interactions. Cell Host Microbe. 2022.30(2):200-215.e12.