Structural Research of Sterol Reductases
Sterol reductases are involved in steroid biosynthesis and include Δ (14)-sterol reductase (C14SR, TM7SF2), Δ (7)-sterol reductase (DHCR7), and Δ (24)-sterol reductase (DHCR24). It can reduce the double bond of cholesterol biosynthesis intermediates. Recently, research has revealed that steroid hormones are essential for stress response, immune system regulation, and reproduction in humans. Therefore, structural analysis of intact membrane sterol reductase allows a molecular understanding of DHCR7 and LBR gene mutations in human congenital diseases.
Structural analysis method of sterol reductase MaSR1
To gain a deeper understanding of sterol reductase, researchers identified crystals of MaSR1 from the methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. NADPH is used to crystallize MaSR1 in space group pp1, and the crystals' diffraction is anisotropic. The structure is determined by single-wavelength anomalous dispersion based on selenium and refined at 2.74 Å resolution. Correct atomic modeling is confirmed by introducing additional selenium anomalous scatterers by selective mutagenesis and preparing platinum derivatives.
Structural analysis of MaSR1
Two MaSR1 molecules are aggregated into a dimer to form an asymmetric unit. The enzyme contains ten transmembrane helices (TM1-10) with the amino and carboxyl termini pointing toward the cytoplasm. Two short antiparallel β-sheet regions are scattered in the cytoplasmic exposure loop, as well as two short α-helices (α1 and α2). Residues 1-23 and 162-176 (part of the loop between TM4 and TM5) are not visible in the electron density map. The binding pocket of NADPH is localized in the c-terminal structural domain (TM6-10), and the cavity of unknown ligand density facing the lipid bilayer is surrounded by TM7 and TM10.
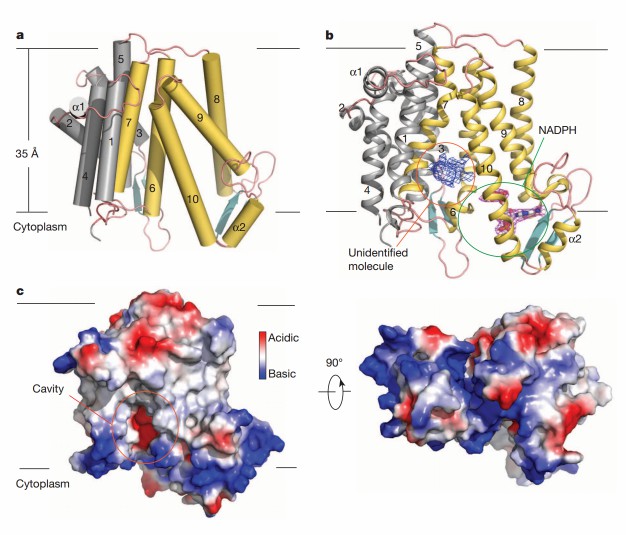 Figure 1. The molecular architecture of MaSR1. (Li X, et al., 2015)
Figure 1. The molecular architecture of MaSR1. (Li X, et al., 2015)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| An integral membrane delta (14)-sterol reductase | Methylotuvimicrobium alcaliphilum 20Z | X-ray diffraction | 2.743 Å | 4QUV |
| Delta (4)-3-ketosteroid 5-beta-reductase in complex with NADP and Testosterone | Homo sapiens | X-ray diffraction | 1.62 Å | 3BUR |
| Delta (4)-3-ketosteroid 5-beta-reductase in complex with NADP and HEPES | Homo sapiens | X-ray diffraction | 1.35 Å | 3BUV |
| Delta (4)-3-ketosteroid 5-beta-reductase in complex with NADP and glycerol | Homo sapiens | X-ray diffraction | 1.79 Å | 3BV7 |
| Delta (4)-3-ketosteroid 5beta-reductase (akr1d1) in complex with progesterone and NADP | Homo sapiens | X-ray diffraction | 2.03 Å | 3COT |
| 5beta-reductase (AKR1D1) in complex with NADPH | Homo sapiens | X-ray diffraction | 2.2 Å | 3CAQ |
| 5beta-reductase (AKR1D1) in complex with NADP+ and 4-androstenedione | Homo sapiens | X-ray diffraction | 2 Å | 3CAS |
| 5beta-reductase (AKR1D1) in complex with NADP+ and 5beta-pregnan-3,20-dione | Homo sapiens | X-ray diffraction | 1.9 Å | 3CAV |
Table 1. Structural research of the sterol reductases.
Creative Biostructure provides high-quality structural biology solutions to support drug discovery, protein engineering, and basic research. Our experienced scientists are well-versed in a variety of techniques such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR).
Whether you need a full range of structural resolution of sterol reductases or other complex proteins, our services will satisfy your specific requirements. We are committed to providing timely and accurate results. Please feel free to contact us with any questions or to discuss your project in more detail.
Reference
- Li X, et al. Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum. Nature. 2015. 517(7532): 104-107.
