Structural Research of Monotopic Glycosyltransferases
Glycosyltransferases (GTs, EC 2.4. xy) are the most diverse class of enzymes present in nature. In eukaryotic cell, the vast majority of glycosyltransferases are located in the endoplasmic reticulum and Golgi apparatus. Except for a few, they are type II membrane proteins with similar domain structures. The molecular mechanism of the glycosylation reaction has long been unclear, the most important reason being the lack of understanding of the three-dimensional structure of glycosyltransferases. the advent of X-ray crystallography has led to great progress in the determination of the crystal structure of GT enzymes. Currently, so many GT families use only two folding methods, forming two superfamilies named GT-A superfamily and GT-B superfamily. These two superfamilies are different in terms of folding methods, active sites and catalytic mechanisms.
The members of the GT-A superfamily all have similar structural characteristics. The two closely connected different domains form a conical shape with a large "pocket" at the top to accommodate the donor and receptor, where the N-terminal domain binds to the NDP sugar donor and the C-terminal domain binds to the receptor. One of the biggest characteristics of the GT-A superfamily is that they all contain a highly conservative "DXD" motif. Someone has proved that this first mock examination motif plays an indispensable role in the catalytic activity of enzymes through site-directed mutation, and the divalent metal cations (usually Mn2+) also play an important role in the catalytic activity. Through their coordination, the "DXD" motif is connected to the phosphate group of NDP sugar donors, in addition, the divalent cation can also play a role in stabilizing the negative charge of the phosphate group and making the chemical bond between nucleotide and sugar fragile and easy to break.
The structural feature of the GT-B ultrasound family is that two similar structural domains are separated by a deep crack, and both domains use a folding method similar to Rossman α/β/α. The C-terminal domain is a typical nucleotide-binding domain (NBD) that binds to NDP sugar donors located in cracks. Unlike the GT-A superfamily, there is no "DXD" motif in the GT-B superfamily, and catalytic reactions also require the participation of metal ions. The folding structure of α/β/α plays a crucial role, and the helical dipole effect of α/β/α can stabilize the negative charge of phosphate groups in the donor, The two α helices also bind to corresponding groups in the donor.
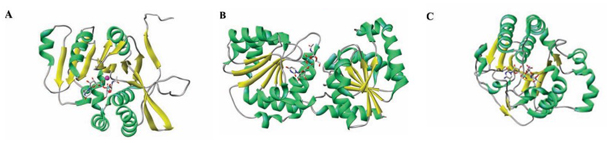 Figure 1. Ribbon diagram of three glycosyltransferases (GTs) representative of the different folds. (BRETON C, et al., 2005)
Figure 1. Ribbon diagram of three glycosyltransferases (GTs) representative of the different folds. (BRETON C, et al., 2005)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Peptidoglycan Glycosyltransferase | Staphylococcus aureus | X-ray diffraction | 2.80 Å | 2OLV |
| PBP1a | Aquifex aeolicus VF5 | X-ray diffraction | 2.10 Å | 2OQO |
| PBP1b | Escherichia coli K-12 | X-ray diffraction | 2.16 Å | 3VMA |
| PBP1b | Escherichia coli | X-ray diffraction | 3.09 Å | 3FWL |
| PBP1b | Escherichia coli K-12 | Cryo-EM analysis | 3.28 Å | 7LQ6 |
| Monofunctional glycosyltransferase WaaA, substrate free | Aquifex aeolicus | X-ray diffraction | 2.00 Å | 2XCI |
| Monofunctional glycosyltransferase WaaA, complex with CMP | Aquifex aeolicus | X-ray diffraction | 2.42 Å | 2XCU |
| Monofunctional glycosyltransferase in complex with Lipid II analog | Staphylococcus aureus subsp. aureus Mu50 | X-ray diffraction | 2.30 Å | 3VMT |
| Monofunctional glycosyltransferase in complex with Lipid II analog, substrate-free protein | Staphylococcus aureus subsp. aureus Mu50 | X-ray diffraction | 2.52 Å | 3VMQ |
| Monofunctional glycosyltransferase in complex with Lipid II analog, in complex with moenomycin | Staphylococcus aureus subsp. aureus Mu50 | X-ray diffraction | 3.69 Å | 3VMR |
| Monofunctional glycosyltransferase in complex with Lipid II analog, in complex with NBD-Lipid II | Staphylococcus aureus subsp. aureus Mu50 | X-ray diffraction | 3.20 Å | 3VMS |
| PglH glycosyltransferase in complex with UDP-galNAc | Campylobacter jejuni | X-ray diffraction | 2.30 Å | 6EJI |
| PglH glycosyltransferase in complex with UDP & synthetic LLO | Campylobacter jejuni | X-ray diffraction | 2.70 Å | 6EJJ |
| PglH glycosyltransferase in complex with UDP-CH2-GalNAc | Campylobacter jejuni | X-ray diffraction | 3.30 Å | 6EJK |
Table 1. Structural Research of Glycosyltransferase.
Creative Biostructure provides customers with membrane protein analysis services based on cryo-electron microscopy technology. This is a powerful imaging technique that can display biomolecules at high resolution. This technology has revolutionized the field of structural biology, allowing researchers to study protein complexes, viruses and other biological macromolecules in their natural environment. Cryo-electron microscopy involves rapidly freezing samples in liquid nitrogen to preserve their structure, and then using electron microscopy to capture images of these samples. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- BRETON C, et al. Structures and mechanisms of glycosyltransferases. Glycobiology, 2005, 16(2).
- LOVERING A L, et al. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science, 2007, 315(5817): 1402–1405.
- YUAN Y, et al. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proceedings of the National Academy of Sciences, 2007, 104(13): 5348–5353.
- SUNG M-T, et al. Crystal structure of the membrane-bound bifunctional transglycosylase pbp1b from escherichia coli. Proceedings of the National Academy of Sciences, 2009, 106(22): 8824–8829.
- SCHMIDT H, et al. Structural and mechanistic analysis of the membrane-embedded glycosyltransferase WAAA required for lipopolysaccharide synthesis. Proceedings of the National Academy of Sciences, 2012, 109(16): 6253–6258.
- HUANG C-Y, et al. Crystal structure of staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism. Proceedings of the National Academy of Sciences, 2012, 109(17): 6496–6501.
- RAMÍREZ A S, et al. Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase. Nature Communications, 2018, 9(1).