Structural Research of Thioesterases
Thioesterases are a crucial class of enzymes found in all living cells that regulate intracellular metabolic pathways through the hydrolysis or synthesis of thioesterase bonds. They are involved in biological processes such as signal transduction and regulation. Structural analysis of Thioesterases can reveal their catalytic mechanism, substrate binding modes, and interactions with other proteins or small molecules. This structural information guides the designing of potential drug targets, engineering enzymes to catalyze specific reactions, and developing applications in agriculture and energy. It also provides scientists with a basis for further research and applications for this class of protein.
Thioesterases are classified into 25 families based on sequence conservation, tertiary and quaternary structure, active site configuration, and substrate specificity. Structurally, these families share a common folding and oligomeric state that binds together essential but often different active site configurations. TE4-15 and TE24-25 families have a hotdog folding domain with a single or double structural domain within the primary structure. This domain can self-associate to form higher-order complexes. These hotdog domain thioesterases are found in both prokaryotes and eukaryotes. Families TE2 and TE16-22 contain α/β hydrolase folds, are predominantly monomeric, and are found in both prokaryotes and eukaryotes. The remaining families form three unrelated folds, NagB (TE1), flavin oxytocin (TE3), and lactamase (TE23). In addition, the presence of structural domain duplications in many TE families suggests gene duplication events. This is where genomes once located in different reading frames are combined to form a single open reading frame containing multiple structural domains.
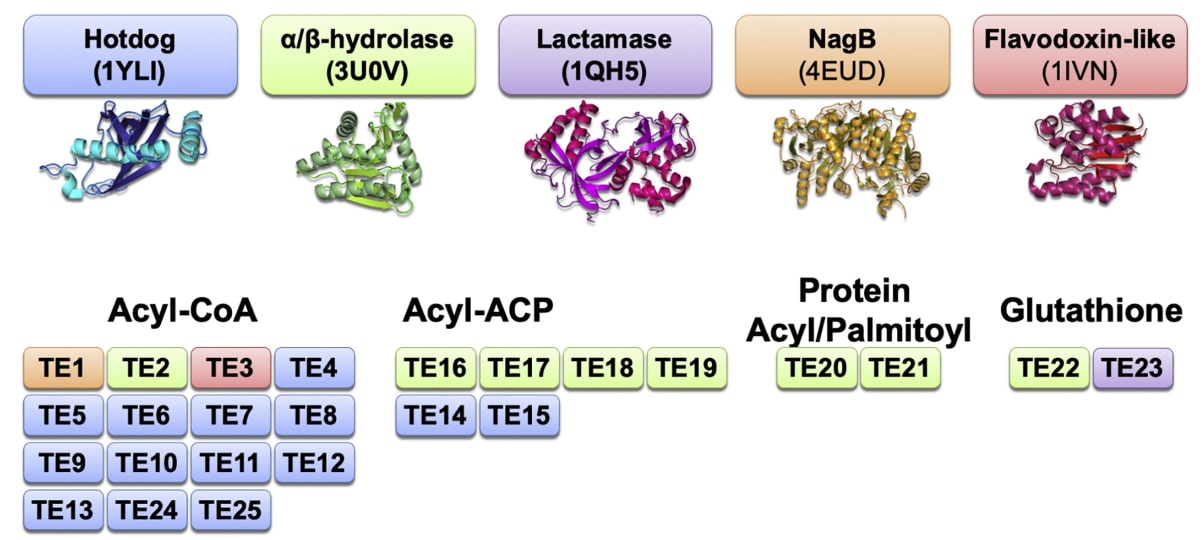 Figure 1. An overview of the domain types present in different thioesterases. (Swarbrick CMD, et al., 2020)
Figure 1. An overview of the domain types present in different thioesterases. (Swarbrick CMD, et al., 2020)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| acyl protein thioesterase 1 (APT 1) bound to palmitic acid: Homo sapiens | Homo sapiens | X-ray diffraction | 2.75 Å | 6QGS |
| APT1 C2S mutant bound to palmitic acid | Homo sapiens | X-ray diffraction | 2.60 Å | 6QGQ |
| APT1 S119A mutant bound to palmitic acid | Homo sapiens | X-ray diffraction | 2.60 Å | 6QGO |
| APT1 bound to 2-Bromopalmitate | Homo sapiens | X-ray diffraction | 2.10 Å | 6QGN |
Table 1. Structural research of thioesterases.
Structural biology methods and techniques play a crucial role in the study of thioesterase structure. X-ray crystallography is the most common method used to resolve protein structures and can be used to determine their high-resolution 3D structures at the atomic level from diffraction images of protein crystals.
Creative Biostructure, a leading service provider in the field of structural biology, offers a wide range of services and solutions related to thioesterase structural analysis. For example, we offer X-ray crystallography, cryo-electron microscopy (Cryo-EM), and nuclear magnetic resonance (NMR) services. In addition, we also provide technical support for protein expression and purification, protein crystallization screening, protein structure modeling and analysis, and much more. For more information, please contact us.
References
- Abrami L, et al. Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains. Nat Chem Biol. 2021; 17(4): 438-447.
- Swarbrick CMD, et al. Structure, function, and regulation of thioesterases. Prog Lipid Res. 2020. 79: 101036.